Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus
- PMID: 18573903
- PMCID: PMC2519419
- DOI: 10.1128/IAI.00208-08
Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus
Abstract
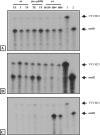
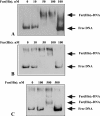
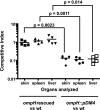
Vibrio vulnificus multiplies rapidly in host tissues under iron-overloaded conditions. To understand the effects of iron in the physiology of this pathogen, we performed a genome-wide transcriptional analysis of V. vulnificus growing at three different iron concentrations, i.e., iron-limiting [Trypticase soy broth with 1.5% NaCl (TSBS) plus ethylenediamine-di-(o-hydroxyphenylacetic) acid (EDDA)], low-iron (1 microg Fe/ml; TSBS), and iron-rich (38 microg Fe/ml; TSBS plus ferric ammonium citrate) concentrations. A few genes were upregulated under the last two conditions, while several genes were expressed differentially under only one of them. A gene upregulated under both conditions encodes the outer membrane porin, OmpH, while others are related to the biosynthesis of amino sugars. An ompH mutant showed sensitivity to sodium dodecyl sulfate (SDS) and polymyxin B and also had a reduced competitive index compared with the wild type in the iron-overloaded mice. Under iron-limiting conditions, two of the TonB systems involved in vulnibactin transport were induced. These genes were essential for virulence in the iron-overloaded mice inoculated subcutaneously, underscoring the importance of active iron transport in infection, even under the high-iron conditions of this animal model. Furthermore, we demonstrated that a RyhB homologue is also essential for virulence in the iron-overloaded mouse. This novel information on the role of genes induced under iron limitation in the iron-overloaded mouse model and the finding of new genes with putative roles in virulence that are expressed only under iron-rich conditions shed light on the many strategies used by this pathogen to multiply rapidly in the susceptible host.
Figures


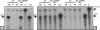


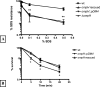

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. - PubMed
-
- Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19185-193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

