Structural and mutational characterization of the catalytic A-module of the mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii
- PMID: 18574239
- PMCID: PMC3259796
- DOI: 10.1074/jbc.M804119200
Structural and mutational characterization of the catalytic A-module of the mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii
Abstract
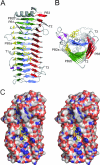
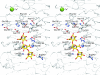
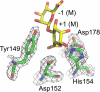
Alginate is a family of linear copolymers of (1-->4)-linked beta-d-mannuronic acid and its C-5 epimer alpha-l-guluronic acid. The polymer is first produced as polymannuronic acid and the guluronic acid residues are then introduced at the polymer level by mannuronan C-5-epimerases. The structure of the catalytic A-module of the Azotobacter vinelandii mannuronan C-5-epimerase AlgE4 has been determined by x-ray crystallography at 2.1-A resolution. AlgE4A folds into a right-handed parallel beta-helix structure originally found in pectate lyase C and subsequently in several polysaccharide lyases and hydrolases. The beta-helix is composed of four parallel beta-sheets, comprising 12 complete turns, and has an amphipathic alpha-helix near the N terminus. The catalytic site is positioned in a positively charged cleft formed by loops extending from the surface encompassing Asp(152), an amino acid previously shown to be important for the reaction. Site-directed mutagenesis further implicates Tyr(149), His(154), and Asp(178) as being essential for activity. Tyr(149) probably acts as the proton acceptor, whereas His(154) is the proton donor in the epimerization reaction.
Figures

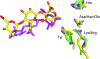

Similar articles
-
Structural and functional characterization of the R-modules in alginate C-5 epimerases AlgE4 and AlgE6 from Azotobacter vinelandii.J Biol Chem. 2014 Nov 7;289(45):31382-96. doi: 10.1074/jbc.M114.567008. Epub 2014 Sep 29. J Biol Chem. 2014. PMID: 25266718 Free PMC article.
-
NMR structure of the R-module: a parallel beta-roll subunit from an Azotobacter vinelandii mannuronan C-5 epimerase.J Biol Chem. 2006 Mar 17;281(11):7350-6. doi: 10.1074/jbc.M510069200. Epub 2006 Jan 3. J Biol Chem. 2006. PMID: 16407237
-
The catalytic activities of the bifunctional Azotobacter vinelandii mannuronan C-5-epimerase and alginate lyase AlgE7 probably originate from the same active site in the enzyme.J Biol Chem. 2001 Aug 24;276(34):31542-50. doi: 10.1074/jbc.M102562200. Epub 2001 Jun 4. J Biol Chem. 2001. PMID: 11390391
-
Mannuronate C-5 epimerases and their use in alginate modification.Essays Biochem. 2023 Apr 18;67(3):615-627. doi: 10.1042/EBC20220151. Essays Biochem. 2023. PMID: 36876890 Review.
-
Alginate-modifying enzymes: biological roles and biotechnological uses.Front Microbiol. 2015 May 27;6:523. doi: 10.3389/fmicb.2015.00523. eCollection 2015. Front Microbiol. 2015. PMID: 26074905 Free PMC article. Review.
Cited by
-
The three-sided right-handed β-helix is a versatile fold for glycan interactions.Glycobiology. 2024 May 26;34(7):cwae037. doi: 10.1093/glycob/cwae037. Glycobiology. 2024. PMID: 38767844 Free PMC article. Review.
-
Structural and functional study of D-glucuronyl C5-epimerase.J Biol Chem. 2015 Feb 20;290(8):4620-4630. doi: 10.1074/jbc.M114.602201. Epub 2015 Jan 7. J Biol Chem. 2015. PMID: 25568314 Free PMC article.
-
Occurrence of L-iduronic acid and putative D-glucuronyl C5-epimerases in prokaryotes.Glycoconj J. 2011 Feb;28(2):57-66. doi: 10.1007/s10719-011-9324-7. Epub 2011 Feb 24. Glycoconj J. 2011. PMID: 21347714 Free PMC article.
-
Structural and functional characterization of the R-modules in alginate C-5 epimerases AlgE4 and AlgE6 from Azotobacter vinelandii.J Biol Chem. 2014 Nov 7;289(45):31382-96. doi: 10.1074/jbc.M114.567008. Epub 2014 Sep 29. J Biol Chem. 2014. PMID: 25266718 Free PMC article.
-
Biosynthesis of the Pseudomonas aeruginosa Extracellular Polysaccharides, Alginate, Pel, and Psl.Front Microbiol. 2011 Aug 22;2:167. doi: 10.3389/fmicb.2011.00167. eCollection 2011. Front Microbiol. 2011. PMID: 21991261 Free PMC article.
References
-
- Fischer, F. G., and Dörfel, H. (1955) Hoppe-Seyler's Z. Physiol. Chem. 302 186–203 - PubMed
-
- Drummond, D. W., Hirst, E. L., and Percival, E. (1962) J. Chem. Soc. pp. 1208–1216
-
- Linker, A., and Jones, R. S. (1964) Nature 204 187–188 - PubMed
-
- Gorin, P. A. J., and Spencer, J. F. T. (1966) Can. J. Chem. 44 993–998
-
- Donati, I., Holtan, S., Mørch, Y. A., Borgogna, M., Dentini, M., and Skjåk-Bræk, G. (2005) Biomacromolecules 6 1031–1040 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

