Functional characterization of fingers subdomain-specific monoclonal antibodies inhibiting the hepatitis C virus RNA-dependent RNA polymerase
- PMID: 18574240
- PMCID: PMC3259772
- DOI: 10.1074/jbc.M803422200
Functional characterization of fingers subdomain-specific monoclonal antibodies inhibiting the hepatitis C virus RNA-dependent RNA polymerase
Abstract
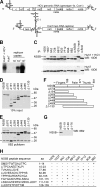
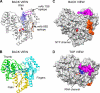
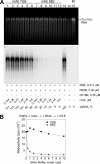
The hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp), encoded by nonstructural protein 5B (NS5B), is absolutely essential for the viral replication. Here we describe the development, characterization, and functional properties of the panel of monoclonal antibodies (mAbs) and specifically describe the mechanism of action of two mAbs inhibiting the NS5B RdRp activity. These mAbs recognize and bind to distinct linear epitopes in the fingers subdomain of NS5B. The mAb 8B2 binds the N-terminal epitope of the NS5B and inhibits both primer-dependent and de novo RNA synthesis. mAb 8B2 selectively inhibits elongation of RNA chains and enhances the RNA template binding by NS5B. In contrast, mAb 7G8 binds the epitope that contains motif G conserved in viral RdRps and inhibits only primer-dependent RNA synthesis by specifically targeting the initiation of RNA synthesis, while not interfering with the binding of template RNA by NS5B. To reveal the importance of the residues of mAb 7G8 epitope for the initiation of RNA synthesis, we performed site-directed mutagenesis and extensively characterized the functionality of the HCV RdRp motif G. Comparison of the mutation effects in both in vitro primer-dependent RdRp assay and cellular transient replication assay suggested that mAb 7G8 epitope amino acid residues are involved in the interaction of template-primer or template with HCV RdRp. The data presented here allowed us to describe the functionality of the epitopes of mAbs 8B2 and 7G8 in the HCV RdRp activity and suggest that the epitopes recognized by these mAbs may be useful targets for antiviral drugs.
Figures
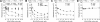



Similar articles
-
Monoclonal antibody recognizing N-terminal epitope of hepatitis C virus nonstructural 5B inhibits viral RNA replication.J Viral Hepat. 2008 Apr;15(4):305-13. doi: 10.1111/j.1365-2893.2007.00945.x. J Viral Hepat. 2008. PMID: 18307593
-
Characterization of monoclonal antibodies that specifically recognize the palm subdomain of hepatitis C virus nonstructural protein 5B polymerase.Virus Res. 2001 Jun;75(2):179-87. doi: 10.1016/s0168-1702(01)00239-8. Virus Res. 2001. PMID: 11325472
-
Interference of HCV replication by cell penetrable human monoclonal scFv specific to NS5B polymerase.MAbs. 2014;6(5):1327-39. doi: 10.4161/mabs.29978. MAbs. 2014. PMID: 25517317 Free PMC article.
-
Recent advances in discovery and development of promising therapeutics against hepatitis C virus NS5B RNA-dependent RNA polymerase.Mini Rev Med Chem. 2005 Dec;5(12):1103-12. doi: 10.2174/138955705774933310. Mini Rev Med Chem. 2005. PMID: 16375756 Review.
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
Cited by
-
RNA Interference-Guided Targeting of Hepatitis C Virus Replication with Antisense Locked Nucleic Acid-Based Oligonucleotides Containing 8-oxo-dG Modifications.PLoS One. 2015 Jun 3;10(6):e0128686. doi: 10.1371/journal.pone.0128686. eCollection 2015. PLoS One. 2015. PMID: 26039055 Free PMC article.
-
Globular domain structure and function of restriction-like-endonuclease LINEs: similarities to eukaryotic splicing factor Prp8.Mob DNA. 2017 Nov 7;8:16. doi: 10.1186/s13100-017-0097-9. eCollection 2017. Mob DNA. 2017. PMID: 29151899 Free PMC article.
-
Intracytoplasmic stable expression of IgG1 antibody targeting NS3 helicase inhibits replication of highly efficient hepatitis C Virus 2a clone.Virol J. 2010 Jun 7;7:118. doi: 10.1186/1743-422X-7-118. Virol J. 2010. PMID: 20529250 Free PMC article.
-
Progress towards recombinant anti-infective antibodies.Recent Pat Antiinfect Drug Discov. 2009 Jan;4(1):1-17. doi: 10.2174/157489109787236319. Recent Pat Antiinfect Drug Discov. 2009. PMID: 19149692 Free PMC article. Review.
-
Dissection of two drug-targeted regions of Hepatitis C virus subtype 4a infecting Egyptian patients.Virus Genes. 2020 Oct;56(5):564-581. doi: 10.1007/s11262-020-01776-y. Epub 2020 Jun 22. Virus Genes. 2020. PMID: 32572756 Free PMC article.
References
-
- Choo, Q. L., Kuo, G., Weiner, A. J., Overby, L. R., Bradley, D. W., and Houghton, M. (1989) Science 244 pa359-362 - PubMed
-
- Hoofnagle, J. H. (2002) Hepatology 36 Suppl. 1, 21-29 - PubMed
-
- Perz, J. P., Farrington, L. A., Pecoraro, C., Hutin, Y. J., and Armstrong, G. L. (2004) 42nd Annual Meeting of the Infectious Diseases Society of America, September 30-October 3, p. 214, Boston, MA
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

