Three consecutive arginines are important for the mycobacterial peptide deformylase enzyme activity
- PMID: 18574247
- PMCID: PMC3259783
- DOI: 10.1074/jbc.M709672200
Three consecutive arginines are important for the mycobacterial peptide deformylase enzyme activity
Abstract
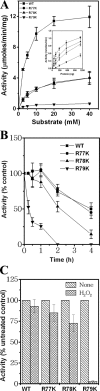
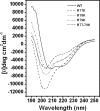
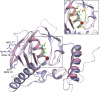
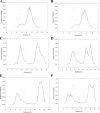
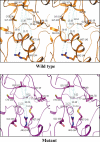
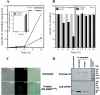
Genes encoding the peptide deformylase enzyme (def) are present in all eubacteria and are involved in the deformylation of the N-formyl group of newly synthesized polypeptides during protein synthesis. We compared the amino acid sequences of this enzyme in different mycobacterial species and found that they are highly conserved (76% homology with 62% identity); however, when this comparison was extended to other eubacterial homologs, it emerged that the mycobacterial proteins have an insertion region containing three consecutive arginine residues (residues 77-79 in Mycobacterium tuberculosis peptide deformylase (mPDF)). Here, we demonstrate that these three arginines are important for the activity of mPDF. Circular dichroism studies of wild-type mPDF and of mPDF containing individual conservative substitutions (R77K, R78K, or R79K) or combined substitutions incorporated into a triple mutant (R77K/R78K/R79K) indicate that such mutations cause mPDF to undergo structural alterations. Molecular modeling of mPDF suggests that the three arginines are distal to the active site. Molecular dynamics simulations of wild-type and mutant mPDF structures indicate that the arginines may be involved in the stabilization of substrate binding pocket residues for their proper interaction with peptide(s). Treatment with 5'-phosphothiorate-modified antisense oligodeoxyribonucleotides directed against different regions of def from M. tuberculosis inhibits growth of Mycobacterium smegmatis in culture. Taken together, these results hold out the possibility of future design of novel mycobacteria-specific PDF inhibitors.
Figures

Similar articles
-
Identification of crucial amino acids of bacterial Peptide deformylases affecting enzymatic activity in response to oxidative stress.J Bacteriol. 2014 Jan;196(1):90-9. doi: 10.1128/JB.00916-13. Epub 2013 Oct 18. J Bacteriol. 2014. PMID: 24142250 Free PMC article.
-
The carboxy-terminal end of the peptide deformylase from Mycobacterium tuberculosis is indispensable for its enzymatic activity.Biochem Biophys Res Commun. 2005 Jul 1;332(2):418-25. doi: 10.1016/j.bbrc.2005.04.142. Biochem Biophys Res Commun. 2005. PMID: 15896710
-
Peptide Deformylase (def) is essential in Mycobacterium smegmatis, but the essentiality is compensated by inactivation of methionine formylation.BMC Microbiol. 2019 Oct 26;19(1):232. doi: 10.1186/s12866-019-1611-7. BMC Microbiol. 2019. PMID: 31655553 Free PMC article.
-
Glycine in the conserved motif III modulates the thermostability and oxidative stress resistance of peptide deformylase in Mycobacterium tuberculosis.FEMS Microbiol Lett. 2011 Jul;320(1):40-7. doi: 10.1111/j.1574-6968.2011.02289.x. Epub 2011 May 16. FEMS Microbiol Lett. 2011. PMID: 21507054
-
Structural and kinetic studies on adenylosuccinate lyase from Mycobacterium smegmatis and Mycobacterium tuberculosis provide new insights on the catalytic residues of the enzyme.FEBS J. 2014 Mar;281(6):1642-58. doi: 10.1111/febs.12730. Epub 2014 Feb 20. FEBS J. 2014. PMID: 24479855
Cited by
-
Amino-terminal extension present in the methionine aminopeptidase type 1c of Mycobacterium tuberculosis is indispensible for its activity.BMC Biochem. 2011 Jul 5;12:35. doi: 10.1186/1471-2091-12-35. BMC Biochem. 2011. PMID: 21729287 Free PMC article.
-
High tolerance to mutations in a Chlamydia trachomatis peptide deformylase loop.World J Biol Chem. 2011 May 26;2(5):90-7. doi: 10.4331/wjbc.v2.i5.90. World J Biol Chem. 2011. PMID: 21666811 Free PMC article.
-
Identification of crucial amino acids of bacterial Peptide deformylases affecting enzymatic activity in response to oxidative stress.J Bacteriol. 2014 Jan;196(1):90-9. doi: 10.1128/JB.00916-13. Epub 2013 Oct 18. J Bacteriol. 2014. PMID: 24142250 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

