Bridging fluorescence microscopy and electron microscopy
- PMID: 18575880
- PMCID: PMC2491700
- DOI: 10.1007/s00418-008-0460-5
Bridging fluorescence microscopy and electron microscopy
Abstract
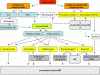
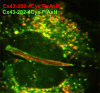
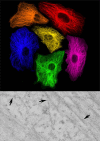
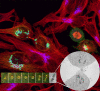
Development of new fluorescent probes and fluorescence microscopes has led to new ways to study cell biology. With the emergence of specialized microscopy units at most universities and research centers, the use of these techniques is well within reach for a broad research community. A major breakthrough in fluorescence microscopy in biology is the ability to follow specific targets on or in living cells, revealing dynamic localization and/or function of target molecules. One of the inherent limitations of fluorescence microscopy is the resolution. Several efforts are undertaken to overcome this limit. The traditional and most well-known way to achieve higher resolution imaging is by electron microscopy. Moreover, electron microscopy reveals organelles, membranes, macromolecules, and thus aids in the understanding of cellular complexity and localization of molecules of interest in relation to other structures. With the new probe development, a solid bridge between fluorescence microscopy and electron microscopy is being built, even leading to correlative imaging. This connection provides several benefits, both scientifically as well as practically. Here, I summarize recent developments in bridging microscopy.
Figures
References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1126/science.1127344', 'is_inner': False, 'url': 'https://doi.org/10.1126/science.1127344'}, {'type': 'PubMed', 'value': '16902090', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/16902090/'}]}
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1126/science.281.5385.2013', 'is_inner': False, 'url': 'https://doi.org/10.1126/science.281.5385.2013'}, {'type': 'PubMed', 'value': '9748157', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/9748157/'}]}
- Bruchez M Jr, Moronne M, Gin P, Weiss S, Alivisatos AP (1998) Semiconductor nanocrystals as fluorescent biological labels. Science 281:2013–2016 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1126/science.281.5385.2016', 'is_inner': False, 'url': 'https://doi.org/10.1126/science.281.5385.2016'}, {'type': 'PubMed', 'value': '9748158', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/9748158/'}]}
- Chan WC, Nie S (1998) Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 281:2016–2018 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/j.copbio.2004.12.003', 'is_inner': False, 'url': 'https://doi.org/10.1016/j.copbio.2004.12.003'}, {'type': 'PubMed', 'value': '15722013', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/15722013/'}]}
- Chen I, Ting AY (2005) Site-specific labeling of proteins with small molecules in live cells. Curr Opin Biotechnol 16:35–40 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/j.tibtech.2005.10.005', 'is_inner': False, 'url': 'https://doi.org/10.1016/j.tibtech.2005.10.005'}, {'type': 'PubMed', 'value': '16269193', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/16269193/'}]}
- Chudakov DM, Lukyanov S, Lukyanov KA (2005) Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol 23:605–613 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

