Off-target effects of siRNA specific for GFP
- PMID: 18577207
- PMCID: PMC2443166
- DOI: 10.1186/1471-2199-9-60
Off-target effects of siRNA specific for GFP
Abstract
Background: Gene knock down by RNAi is a highly effective approach to silence gene expression in experimental as well as therapeutic settings. However, this widely used methodology entails serious pitfalls, especially concerning specificity of the RNAi molecules.
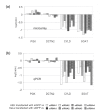
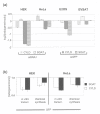
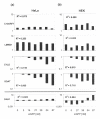
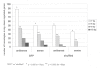
Results: We tested the most widely used control siRNA directed against GFP for off-target effects and found that it deregulates in addition to GFP a set of endogenous target genes. The off-target effects were dependent on the amount of GFP siRNA transfected and were detected in a variety of cell lines. Since the respective siRNA molecule specific for GFP is widely used as negative control for RNAi experiments, we studied the complete set of off-target genes of this molecule by genome-wide expression profiling. The detected modulated mRNAs had target sequences homologous to the siRNA as small as 8 basepairs in size. However, we found no restriction of sequence homology to 3'UTR of target genes.
Conclusion: We can show that even siRNAs without a physiological target have sequence-specific off-target effects in mammalian cells. Furthermore, our analysis defines the off-target genes affected by the siRNA that is commonly used as negative control and directed against GFP. Since off-target effects can hardly be avoided, the best strategy is to identify false positives and exclude them from the results. To this end, we provide the set of false positive genes deregulated by the commonly used GFP siRNA as a reference resource for future siRNA experiments.
Figures
References
-
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

