Genomic and proteomic analysis of the Alkali-Tolerance Response (AlTR) in Listeria monocytogenes 10403S
- PMID: 18577215
- PMCID: PMC2443805
- DOI: 10.1186/1471-2180-8-102
Genomic and proteomic analysis of the Alkali-Tolerance Response (AlTR) in Listeria monocytogenes 10403S
Abstract
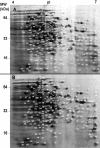
Background: Information regarding the Alkali-Tolerance Response (AlTR) in Listeria monocytogenes is very limited. Treatment of alkali-adapted cells with the protein synthesis inhibitor chloramphenicol has revealed that the AlTR is at least partially protein-dependent. In order to gain a more comprehensive perspective on the physiology and regulation of the AlTR, we compared differential gene expression and protein content of cells adapted at pH 9.5 and un-adapted cells (pH 7.0) using complementary DNA (cDNA) microarray and two-dimensional (2D) gel electrophoresis, (combined with mass spectrometry) respectively.
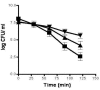
Results: In this study, L. monocytogenes was shown to exhibit a significant AlTR following a 1-h exposure to mild alkali (pH 9.5), which is capable of protecting cells from subsequent lethal alkali stress (pH 12.0). Adaptive intracellular gene expression involved genes that are associated with virulence, the general stress response, cell division, and changes in cell wall structure and included many genes with unknown functions. The observed variability between results of cDNA arrays and 2D gel electrophoresis may be accounted for by posttranslational modifications. Interestingly, several alkali induced genes/proteins can provide a cross protective overlap to other types of stresses.
Conclusion: Alkali pH provides therefore L. monocytogenes with nonspecific multiple-stress resistance that may be vital for survival in the human gastrointestinal tract as well as within food processing systems where alkali conditions prevail. This study showed strong evidence that the AlTR in L. monocytogenes functions as to minimize excess alkalisation and energy expenditures while mobilizing available carbon sources.
Figures
Similar articles
-
Transcriptome analysis of alkali shock and alkali adaptation in Listeria monocytogenes 10403S.Foodborne Pathog Dis. 2010 Oct;7(10):1147-57. doi: 10.1089/fpd.2009.0501. Foodborne Pathog Dis. 2010. PMID: 20677981 Free PMC article.
-
Carbon starvation survival of Listeria monocytogenes in planktonic state and in biofilm: a proteomic study.Proteomics. 2003 Oct;3(10):2052-64. doi: 10.1002/pmic.200300538. Proteomics. 2003. PMID: 14625868
-
Intracellular gene expression profile of Listeria monocytogenes.Infect Immun. 2006 Feb;74(2):1323-38. doi: 10.1128/IAI.74.2.1323-1338.2006. Infect Immun. 2006. PMID: 16428782 Free PMC article.
-
Proteomic analyses of a Listeria monocytogenes mutant lacking sigmaB identify new components of the sigmaB regulon and highlight a role for sigmaB in the utilization of glycerol.Appl Environ Microbiol. 2008 Feb;74(3):594-604. doi: 10.1128/AEM.01921-07. Epub 2007 Dec 7. Appl Environ Microbiol. 2008. PMID: 18065622 Free PMC article.
-
The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics.J Appl Microbiol. 2016 Feb;120(2):251-65. doi: 10.1111/jam.12989. Epub 2015 Dec 28. J Appl Microbiol. 2016. PMID: 26509460 Review.
Cited by
-
Biological Characteristics of Listeria monocytogenes Following Deletion of TatD-like Protein Gene.Curr Microbiol. 2023 Feb 28;80(4):118. doi: 10.1007/s00284-023-03229-9. Curr Microbiol. 2023. PMID: 36853439
-
Dephosphorylation and ion binding in prokaryotic calcium transport.Sci Adv. 2024 Oct 11;10(41):eadp2916. doi: 10.1126/sciadv.adp2916. Epub 2024 Oct 11. Sci Adv. 2024. PMID: 39908574 Free PMC article.
-
Residual Variation Intolerance Score Detects Loci Under Selection in Neuroinvasive Listeria monocytogenes.Front Microbiol. 2019 Nov 26;10:2702. doi: 10.3389/fmicb.2019.02702. eCollection 2019. Front Microbiol. 2019. PMID: 31849867 Free PMC article.
-
Multi-species integrative biclustering.Genome Biol. 2010;11(9):R96. doi: 10.1186/gb-2010-11-9-r96. Epub 2010 Sep 29. Genome Biol. 2010. PMID: 20920250 Free PMC article.
-
Transcriptome analysis of alkali shock and alkali adaptation in Listeria monocytogenes 10403S.Foodborne Pathog Dis. 2010 Oct;7(10):1147-57. doi: 10.1089/fpd.2009.0501. Foodborne Pathog Dis. 2010. PMID: 20677981 Free PMC article.
References
-
- Vasseur C, Rigaud N, Hebraud M, Labadie J. Combined effects of NaCl, NaOH, and biocides (monolaurin or lauric acid) on inactivation of Listeria monocytogenes and Pseudomonas spp. J Food Prot. 2001;64:1442–1445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

