Mass spectrometry analysis of proteome-wide proteolytic post-translational degradation of proteins
- PMID: 18578501
- PMCID: PMC2716136
- DOI: 10.1021/ac800077w
Mass spectrometry analysis of proteome-wide proteolytic post-translational degradation of proteins
Abstract
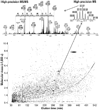
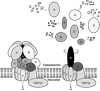
Protein proteolytic degradation is an essential component to proper cell function and its life cycle. Here, we study the protein degradation in yeast Saccharomyces cerevisiae cells on a proteome-wide scale by detection of the intermediate peptides produced from the intracellular degradation of proteins using sequencing-based tandem mass spectrometry. By tracing the detected approximately 1100 peptides and their approximately 200 protein-substrate origins we obtain evidence for new insights into the proteome-wide protein-selective degradation in yeast cells. This evidence shows that the yeast cytoplasm is the largest pool for the degradation of proteins with both biochemical and geometric specificities, whereas the yeast nucleus seems to be a proteolysis-inert organelle under the condition studied. Yeast V-ATPase subunits appear to be degraded during their disassembly, and yeast mitochondrial proteins functioning as precursors, transport carriers, and gates are preferentially degraded. Ubiquitylation may be unnecessary for the proteasomal degradation of yeast cytoplasmic regulatory and enzyme proteins according to our observations. This study shows that the intracellular peptides are informational targets for directly probing the protein degradation-involved molecular mechanisms and cell biology processes.
Figures




References
-
- Goldberg AL. Protein degradation and protection against misfolded or damaged peoteins. Nature. 2003;426:895–899. - PubMed
-
- Demartino GN, Gillette TG. Proteasomes: Machines for all reasons. Cell. 2007;129:659–662. - PubMed
-
- Goldberg AL, Rock KL. Proteolysis, proteasomes and antigen presentation. Nature. 1992;357:375–379. - PubMed
-
- Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigram of a self-compartmentalizing protease. Cell. 1998;92:367–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

