Mutants defective in Rad1-Rad10-Slx4 exhibit a unique pattern of viability during mating-type switching in Saccharomyces cerevisiae
- PMID: 18579504
- PMCID: PMC2516060
- DOI: 10.1534/genetics.108.090654
Mutants defective in Rad1-Rad10-Slx4 exhibit a unique pattern of viability during mating-type switching in Saccharomyces cerevisiae
Abstract
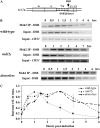
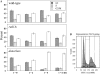
Efficient repair of DNA double-strand breaks (DSBs) requires the coordination of checkpoint signaling and enzymatic repair functions. To study these processes during gene conversion at a single chromosomal break, we monitored mating-type switching in Saccharomyces cerevisiae strains defective in the Rad1-Rad10-Slx4 complex. Rad1-Rad10 is a structure-specific endonuclease that removes 3' nonhomologous single-stranded ends that are generated during many recombination events. Slx4 is a known target of the DNA damage response that forms a complex with Rad1-Rad10 and is critical for 3'-end processing during repair of DSBs by single-strand annealing. We found that mutants lacking an intact Rad1-Rad10-Slx4 complex displayed RAD9- and MAD2-dependent cell cycle delays and decreased viability during mating-type switching. In particular, these mutants exhibited a unique pattern of dead and switched daughter cells arising from the same DSB-containing cell. Furthermore, we observed that mutations in post-replicative lesion bypass factors (mms2Delta, mph1Delta) resulted in decreased viability during mating-type switching and conferred shorter cell cycle delays in rad1Delta mutants. We conclude that Rad1-Rad10-Slx4 promotes efficient repair during gene conversion events involving a single 3' nonhomologous tail and propose that the rad1Delta and slx4Delta mutant phenotypes result from inefficient repair of a lesion at the MAT locus that is bypassed by replication-mediated repair.
Figures



References
-
- Bardwell, A., L. Bardwell, A. Tomkinson and E. Friedberg, 1994. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science 265 2082–2085. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

