Increased mobility of major histocompatibility complex I-peptide complexes decreases the sensitivity of antigen recognition
- PMID: 18579518
- PMCID: PMC3259769
- DOI: 10.1074/jbc.M803549200
Increased mobility of major histocompatibility complex I-peptide complexes decreases the sensitivity of antigen recognition
Abstract
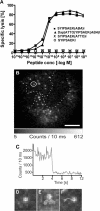
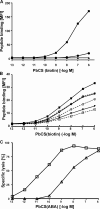
CD8(+) cytotoxic T lymphocytes (CTL) can recognize and kill target cells expressing only a few cognate major histocompatibility complex (MHC) I-peptide complexes. This high sensitivity requires efficient scanning of a vast number of highly diverse MHC I-peptide complexes by the T cell receptor in the contact site of transient conjugates formed mainly by nonspecific interactions of ICAM-1 and LFA-1. Tracking of single H-2K(d) molecules loaded with fluorescent peptides on target cells and nascent conjugates with CTL showed dynamic transitions between states of free diffusion and immobility. The immobilizations were explained by association of MHC I-peptide complexes with ICAM-1 and strongly increased their local concentration in cell adhesion sites and hence their scanning by T cell receptor. In nascent immunological synapses cognate complexes became immobile, whereas noncognate ones diffused out again. Interfering with this mobility modulation-based concentration and sorting of MHC I-peptide complexes strongly impaired the sensitivity of antigen recognition by CTL, demonstrating that it constitutes a new basic aspect of antigen presentation by MHC I molecules.
Figures



Similar articles
-
Recognition of xeno-(HLA, SLA) major histocompatibility complex antigens by mouse cytotoxic T cells is not H-2 restricted: a study with transgenic mice.Proc Natl Acad Sci U S A. 1989 Jan;86(2):617-20. doi: 10.1073/pnas.86.2.617. Proc Natl Acad Sci U S A. 1989. PMID: 2783490 Free PMC article.
-
Endogenous presentation of a nascent antigenic epitope to CD8+ CTL is more efficient than exogenous presentation.Immunol Cell Biol. 1996 Oct;74(5):394-400. doi: 10.1038/icb.1996.68. Immunol Cell Biol. 1996. PMID: 8912001
-
CD8 T cells, like CD4 T cells, are triggered by multivalent engagement of TCRs by MHC-peptide ligands but not by monovalent engagement.J Immunol. 2006 Feb 1;176(3):1498-505. doi: 10.4049/jimmunol.176.3.1498. J Immunol. 2006. PMID: 16424178
-
The diversity of antigen-specific TCR repertoires reflects the relative complexity of epitopes recognized.Hum Immunol. 1997 May;54(2):117-28. doi: 10.1016/s0198-8859(97)00082-7. Hum Immunol. 1997. PMID: 9297530 Review.
-
Interplay between the TCR/CD3 complex and CD4 or CD8 in the activation of cytotoxic T lymphocytes.Immunol Rev. 1989 Jun;109:119-41. doi: 10.1111/j.1600-065x.1989.tb00022.x. Immunol Rev. 1989. PMID: 2527803 Review.
Cited by
-
Co-receptors and recognition of self at the immunological synapse.Curr Top Microbiol Immunol. 2010;340:171-89. doi: 10.1007/978-3-642-03858-7_9. Curr Top Microbiol Immunol. 2010. PMID: 19960314 Free PMC article. Review.
-
MHC molecules lead many lives. Workshop on MHC Class I Molecules at the interface between Biology & Medicine.EMBO Rep. 2009 Jan;10(1):30-4. doi: 10.1038/embor.2008.215. Epub 2008 Dec 5. EMBO Rep. 2009. PMID: 19057577 Free PMC article. No abstract available.
-
A role for rebinding in rapid and reliable T cell responses to antigen.PLoS Comput Biol. 2009 Nov;5(11):e1000578. doi: 10.1371/journal.pcbi.1000578. Epub 2009 Nov 26. PLoS Comput Biol. 2009. PMID: 19956745 Free PMC article.
-
Analysis of molecular diffusion by first-passage time variance identifies the size of confinement zones.Biophys J. 2011 Mar 16;100(6):1463-72. doi: 10.1016/j.bpj.2011.01.064. Biophys J. 2011. PMID: 21402028 Free PMC article.
-
Surface plasmon resonance for high-throughput ligand screening of membrane-bound proteins.Biotechnol J. 2009 Nov;4(11):1542-58. doi: 10.1002/biot.200900195. Biotechnol J. 2009. PMID: 19918786 Free PMC article. Review.
References
-
- Sykulev, Y., Joo, M., Vturina, I., Tsomides, T. J., and Eisen, H. N. (1996) Immunity 4 565-571 - PubMed
-
- Purbhoo, M. A., Irvine, D. J., Huppa, J. B., and Davis, M. M. (2004) Nat. Immunol. 5 524-530 - PubMed
-
- Marwali, M. R., MacLeod, M. A., Muzia, D. N., and Takei, T. (2004) J. Immunol. 173 2960-2967 - PubMed
-
- Suzuki, J., Yamasaki, S., Wu, J., Koretzky, G. J., and Saito, T. (2007) Blood 109 168-175 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

