Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses
- PMID: 18579717
- PMCID: PMC2519498
- DOI: 10.1128/JCM.00192-08
Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses
Abstract
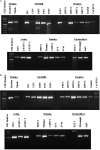
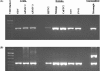
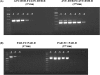
We have developed a set of reverse transcription-PCR assays for the detection and identification of known and novel paramyxoviruses in clinical specimens. Primers were designed from the conserved motifs of the polymerase pol gene sequences to detect members of the Paramyxovirinae or Pneumovirinae subfamily or groups of genera within the Paramyxovirinae subfamily. The consensus-degenerate hybrid oligonucleotide primer design and seminested or nested PCR assay design were used to enhance the breadth of reactivity and sensitivity of the respective assays. Using expressed RNA and 10-fold dilution series of virus-infected tissue culture isolates from different members of the family or genera, these assays were able to detect on average between 100 and 500 copies of template RNA. The assays were specific to the respective group of genera or subfamily viruses. This set of primers enhances our ability to look for novel viruses in outbreaks and diseases of unknown etiology.
Figures




References
-
- Anonymous. 1999. Outbreak of Hendra-like virus—Malaysia and Singapore, 1998-1999. MMWR Morb. Mortal. Wkly. Rep. 48265-269. - PubMed
-
- Kahn, J. S. 2003. Human metapneumovirus: a newly emerging respiratory pathogen. Curr. Opin. Infect. Dis. 16255-258. - PubMed
-
- Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 3481953-1966. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

