Identification of the TFII-I family target genes in the vertebrate genome
- PMID: 18579769
- PMCID: PMC2449355
- DOI: 10.1073/pnas.0803051105
Identification of the TFII-I family target genes in the vertebrate genome
Abstract
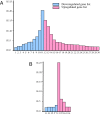
GTF2I and GTF2IRD1 encode members of the TFII-I transcription factor family and are prime candidates in the Williams syndrome, a complex neurodevelopmental disorder. Our previous expression microarray studies implicated TFII-I proteins in the regulation of a number of genes critical in various aspects of cell physiology. Here, we combined bioinformatics and microarray results to identify TFII-I downstream targets in the vertebrate genome. These results were validated by chromatin immunoprecipitation and siRNA analysis. The collected evidence revealed the complexity of TFII-I-mediated processes that involve distinct regulatory networks. Altogether, these results lead to a better understanding of specific molecular events, some of which may be responsible for the Williams syndrome phenotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bayarsaihan D, et al. Genomic organization of the genes Gtf2ird1, Gtf2i, and Ncf1 at the mouse chromosome 5 region syntenic to the human chromosome 7q11.23 Williams syndrome critical region. Genomics. 2002;79:137–143. - PubMed
-
- Polly P, et al. hMusTRD1α1 represses MEF2 activation of the troponin I slow enhancer. J Biol Chem. 2003;278:36603–36610. - PubMed
-
- Vullhorst D, Buonanno A. Characterization of general transcription factor 3, a transcription factor involved in slow muscle-specific gene expression. J Biol Chem. 2003;278:8370–8379. - PubMed
-
- Vullhorst D, Buonanno A. Multiple GTF2I-like repeats of general transcription factor 3 exhibit DNA-binding properties: Evidence for a common origin as a sequence-specific DNA interaction module. J Biol Chem. 2005;280:31722–31731. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

