Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA
- PMID: 18579868
- PMCID: PMC2491469
- DOI: 10.1261/rna.1100708
Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA
Abstract
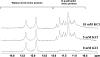
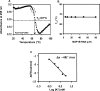
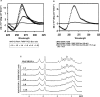
Fragile X mental retardation syndrome, the most common form of inherited mental retardation, is caused by the absence of the fragile X mental retardation protein (FMRP). FMRP has been shown to use its arginine-glycine-glycine (RGG) box to bind to a subset of RNA targets that form a G quadruplex structure. We performed a detailed analysis of the interactions between the FMRP RGG box and the microtubule associated protein 1B (MAP1B) mRNA, a relevant in vivo FMRP target. We show that MAP1B RNA forms an intramolecular G quadruplex structure, which is bound with high affinity and specificity by the FMRP RGG box. We determined that hydrophobic interactions are important in the FMRP RGG box-MAP1B RNA association, with minor contributions from electrostatic interactions. Our findings that at low protein:RNA ratios the RNA G quadruplex structure is slightly stabilized, whereas at high ratios is unfolded, suggest a mechanism by which the FMRP concentration variation in response to a neurotransmitter stimulation event could act as a regulatory switch for the protein function, from translation repressor to translation activator.
Figures
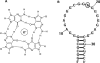



Similar articles
-
Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA.Biochemistry. 2006 Jul 11;45(27):8319-30. doi: 10.1021/bi060209a. Biochemistry. 2006. PMID: 16819831
-
Biophysical characterization of G-quadruplex forming FMR1 mRNA and of its interactions with different fragile X mental retardation protein isoforms.RNA. 2014 Jan;20(1):103-14. doi: 10.1261/rna.041442.113. Epub 2013 Nov 18. RNA. 2014. PMID: 24249225 Free PMC article.
-
Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family.Nucleic Acids Res. 2007;35(16):5379-92. doi: 10.1093/nar/gkm581. Epub 2007 Aug 9. Nucleic Acids Res. 2007. PMID: 17693432 Free PMC article.
-
The fragile X mental retardation protein, FMRP, recognizes G-quartets.Ment Retard Dev Disabil Res Rev. 2004;10(1):49-52. doi: 10.1002/mrdd.20008. Ment Retard Dev Disabil Res Rev. 2004. PMID: 14994288 Review.
-
Identification of messenger RNAs and microRNAs associated with fragile X mental retardation protein.Methods Mol Biol. 2006;342:267-76. doi: 10.1385/1-59745-123-1:267. Methods Mol Biol. 2006. PMID: 16957381 Review.
Cited by
-
FMRP interacts with G-quadruplex structures in the 3'-UTR of its dendritic target Shank1 mRNA.RNA Biol. 2014;11(11):1364-74. doi: 10.1080/15476286.2014.996464. RNA Biol. 2014. PMID: 25692235 Free PMC article.
-
Translational repression of the disintegrin and metalloprotease ADAM10 by a stable G-quadruplex secondary structure in its 5'-untranslated region.J Biol Chem. 2011 Dec 30;286(52):45063-72. doi: 10.1074/jbc.M111.296921. Epub 2011 Nov 7. J Biol Chem. 2011. PMID: 22065584 Free PMC article.
-
New X-chromosomal interactors of dFMRP regulate axonal and synaptic morphology of brain neurons in Drosophila melanogaster.Biotechnol Biotechnol Equip. 2014 Jul 4;28(4):697-709. doi: 10.1080/13102818.2014.937897. Epub 2014 Oct 23. Biotechnol Biotechnol Equip. 2014. PMID: 26740770 Free PMC article.
-
Therapeutic Strategies in Fragile X Syndrome: From Bench to Bedside and Back.Neurotherapeutics. 2015 Jul;12(3):584-608. doi: 10.1007/s13311-015-0355-9. Neurotherapeutics. 2015. PMID: 25986746 Free PMC article. Review.
-
From FMRP function to potential therapies for fragile X syndrome.Neurochem Res. 2014 Jun;39(6):1016-31. doi: 10.1007/s11064-013-1229-3. Epub 2013 Dec 18. Neurochem Res. 2014. PMID: 24346713 Free PMC article. Review.
References
-
- Ambrus A., Chen D., Dai J., Jones R.A., Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. - PubMed
-
- Antar L.N., Bassell G.J. Sunrise at the synapse: The FMRP mRNP shaping the synaptic interface. Neuron. 2003;37:555–558. - PubMed
-
- Antar L.N., Dictenberg J.B., Plociniak M., Afroz R., Bassell G.J. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. - PubMed
-
- Baumann C., Otridge J., Gollnick P. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J. Biol. Chem. 1996;271:12269–12274. - PubMed
-
- Becktel W.J., Schellman J.A. Protein stability curves. Biopolymers. 1987;26:1859–1877. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
