Protein-protein interaction detection in vitro and in cells by proximity biotinylation
- PMID: 18582056
- PMCID: PMC2635094
- DOI: 10.1021/ja801445p
Protein-protein interaction detection in vitro and in cells by proximity biotinylation
Abstract
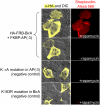
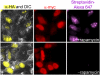
We report a new method for detection of protein-protein interactions in vitro and in cells. One protein partner is fused to Escherichia coli biotin ligase (BirA), while the other protein partner is fused to BirA's "acceptor peptide" (AP) substrate. If the two proteins interact, BirA will catalyze site-specific biotinylation of AP, which can be detected by streptavidin staining. To minimize nonspecific signals, we engineered the AP sequence to reduce its intrinsic affinity for BirA. The rapamycin-controlled interaction between FKBP and FRB proteins could be detected in vitro and in cells with a signal to background ratio as high as 28. We also extended the method to imaging of the phosphorylation-dependent interaction between Cdc25C phosphatase and 14-3-3epsilon phosphoserine/threonine binding protein. Protein-protein interaction detection by proximity biotinylation has the advantages of low background, high sensitivity, small AP tag size, and good spatial resolution in cells.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

