Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis
- PMID: 18583528
- PMCID: PMC2492618
- DOI: 10.1104/pp.108.123802
Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis
Abstract
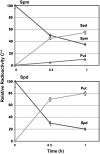
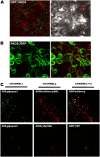
In contrast to animals, where polyamine (PA) catabolism efficiently converts spermine (Spm) to putrescine (Put), plants have been considered to possess a PA catabolic pathway producing 1,3-diaminopropane, Delta(1)-pyrroline, the corresponding aldehyde, and hydrogen peroxide but unable to back-convert Spm to Put. Arabidopsis (Arabidopsis thaliana) genome contains at least five putative PA oxidase (PAO) members with yet-unknown localization and physiological role(s). AtPAO1 was recently identified as an enzyme similar to the mammalian Spm oxidase, which converts Spm to spermidine (Spd). In this work, we have performed in silico analysis of the five Arabidopsis genes and have identified PAO3 (AtPAO3) as a nontypical PAO, in terms of homology, compared to other known PAOs. We have expressed the gene AtPAO3 and have purified a protein corresponding to it using the inducible heterologous expression system of Escherichia coli. AtPAO3 catalyzed the sequential conversion/oxidation of Spm to Spd, and of Spd to Put, thus exhibiting functional homology to the mammalian PAOs. The best substrate for this pathway was Spd, whereas the N(1)-acetyl-derivatives of Spm and Spd were oxidized less efficiently. On the other hand, no activity was detected when diamines (agmatine, cadaverine, and Put) were used as substrates. Moreover, although AtPAO3 does not exhibit significant similarity to the other known PAOs, it is efficiently inhibited by guazatine, a potent PAO inhibitor. AtPAO3 contains a peroxisomal targeting motif at the C terminus, and it targets green fluorescence protein to peroxisomes when fused at the N terminus but not at the C terminus. These results reveal that AtPAO3 is a peroxisomal protein and that the C terminus of the protein contains the sorting information. The overall data reinforce the view that plants and mammals possess a similar PA oxidation system, concerning both the subcellular localization and the mode of its action.
Figures




References
-
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92 773–784 - PubMed
-
- Amendola R, Bellini A, Cervelli M, Degan P, Marcocci L, Martini F, Mariottini P (2005) Direct oxidative DNA damage, apoptosis and radio sensitivity by spermine oxidase activities in mouse neuroblastoma cells. Biochim Biophys Acta 1755 15–24 - PubMed
-
- Angelini R, Federico R, Bonfante P (1995) Maize polyamine oxidase: antibody production and ultrastructural localization. J Plant Physiol 145 686–692
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

