The PRA1 gene family in Arabidopsis
- PMID: 18583532
- PMCID: PMC2492607
- DOI: 10.1104/pp.108.122226
The PRA1 gene family in Arabidopsis
Abstract
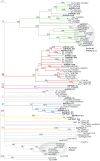
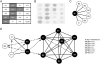
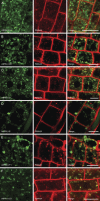
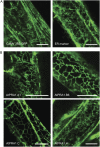
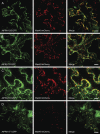
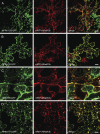
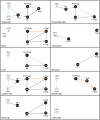
Prenylated Rab acceptor 1 (PRA1) domain proteins are small transmembrane proteins that regulate vesicle trafficking as receptors of Rab GTPases and the vacuolar soluble N-ethylmaleimide-sensitive factor attachment receptor protein VAMP2. However, little is known about PRA1 family members in plants. Sequence analysis revealed that higher plants, compared with animals and primitive plants, possess an expanded family of PRA1 domain-containing proteins. The Arabidopsis (Arabidopsis thaliana) PRA1 (AtPRA1) proteins were found to homodimerize and heterodimerize in a manner corresponding to their phylogenetic distribution. Different AtPRA1 family members displayed distinct expression patterns, with a preference for vascular cells and expanding or developing tissues. AtPRA1 genes were significantly coexpressed with Rab GTPases and genes encoding vesicle transport proteins, suggesting an involvement in the vesicle trafficking process similar to that of their animal counterparts. Correspondingly, AtPRA1 proteins were localized in the endoplasmic reticulum, Golgi apparatus, and endosomes/prevacuolar compartments, hinting at a function in both secretory and endocytic intracellular trafficking pathways. Taken together, our data reveal a high functional diversity of AtPRA1 proteins, probably dealing with the various demands of the complex trafficking system.
Figures

References
-
- Abdul-Ghani M, Gougeon P-Y, Prosser DC, Da-Silva LF, Ngsee JK (2001) PRA isoforms are targeted to distinct membrane compartments. J Biol Chem 276 6225–6233 - PubMed
-
- Akiduki S, Ochiishi T, Ikemoto MJ (2007) Neural localization of addicsin in mouse brain. Neurosci Lett 426 149–154 - PubMed
-
- Aoki K, Ogata Y, Shibata D (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48 381–390 - PubMed
-
- Assaad FF (2001) Of weeds and men: what genomes teach us about plant cell biology. Curr Opin Plant Biol 4 478–487 - PubMed
-
- Bassham DC, Raikhel NV (2000) Unique features of the plant vacuolar sorting machinery. Curr Opin Cell Biol 12 491–495 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

