Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways
- PMID: 18583645
- PMCID: PMC2556274
- DOI: 10.1101/gr.077404.108
Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways
Abstract
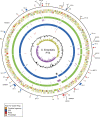
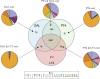
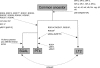
We have determined the complete genome sequences of a host-promiscuous Salmonella enterica serovar Enteritidis PT4 isolate P125109 and a chicken-restricted Salmonella enterica serovar Gallinarum isolate 287/91. Genome comparisons between these and other Salmonella isolates indicate that S. Gallinarum 287/91 is a recently evolved descendent of S. Enteritidis. Significantly, the genome of S. Gallinarum has undergone extensive degradation through deletion and pseudogene formation. Comparison of the pseudogenes in S. Gallinarum with those identified previously in other host-adapted bacteria reveals the loss of many common functional traits and provides insights into possible mechanisms of host and tissue adaptation. We propose that experimental analysis in chickens and mice of S. Enteritidis-harboring mutations in functional homologs of the pseudogenes present in S. Gallinarum could provide an experimentally tractable route toward unraveling the genetic basis of host adaptation in S. enterica.
Figures



Similar articles
-
Genomic comparison between Salmonella Gallinarum and Pullorum: differential pseudogene formation under common host restriction.PLoS One. 2013;8(3):e59427. doi: 10.1371/journal.pone.0059427. Epub 2013 Mar 15. PLoS One. 2013. PMID: 23555032 Free PMC article.
-
Host-specificity of Salmonella enterica serovar Gallinarum: insights from comparative genomics.Infect Genet Evol. 2009 Jul;9(4):468-73. doi: 10.1016/j.meegid.2009.01.004. Epub 2009 Jan 19. Infect Genet Evol. 2009. PMID: 19454277
-
Molecular evolution of Salmonella enterica subsp. enterica serovar Gallinarum biovar Gallinarum in the field.Vet Microbiol. 2019 Aug;235:63-70. doi: 10.1016/j.vetmic.2019.05.019. Epub 2019 May 24. Vet Microbiol. 2019. PMID: 31282380
-
The chicken, the egg and Salmonella enteritidis.Environ Microbiol. 2001 Jul;3(7):421-30. doi: 10.1046/j.1462-2920.2001.00213.x. Environ Microbiol. 2001. PMID: 11553232 Review.
-
Salmonella pathogenicity and host adaptation in chicken-associated serovars.Microbiol Mol Biol Rev. 2013 Dec;77(4):582-607. doi: 10.1128/MMBR.00015-13. Microbiol Mol Biol Rev. 2013. PMID: 24296573 Free PMC article. Review.
Cited by
-
Enteric Fever Diagnosis: Current Challenges and Future Directions.Pathogens. 2021 Apr 1;10(4):410. doi: 10.3390/pathogens10040410. Pathogens. 2021. PMID: 33915749 Free PMC article. Review.
-
Subtyping of Salmonella enterica Subspecies I Using Single-Nucleotide Polymorphisms in Adenylate Cyclase.Foodborne Pathog Dis. 2016 Jul;13(7):350-62. doi: 10.1089/fpd.2015.2088. Epub 2016 Apr 1. Foodborne Pathog Dis. 2016. PMID: 27035032 Free PMC article.
-
Genome sequence of the invasive Salmonella enterica subsp. enterica serotype enteritidis strain LA5.J Bacteriol. 2012 May;194(9):2387-8. doi: 10.1128/JB.00256-12. J Bacteriol. 2012. PMID: 22493198 Free PMC article.
-
Salmonella enterica: survival, colonization, and virulence differences among serovars.ScientificWorldJournal. 2015;2015:520179. doi: 10.1155/2015/520179. Epub 2015 Jan 13. ScientificWorldJournal. 2015. PMID: 25664339 Free PMC article.
-
Evolution of Salmonella enterica virulence via point mutations in the fimbrial adhesin.PLoS Pathog. 2012;8(6):e1002733. doi: 10.1371/journal.ppat.1002733. Epub 2012 Jun 7. PLoS Pathog. 2012. PMID: 22685400 Free PMC article.
References
-
- Barrow P. Experimental infection of chickens with Salmonella enteritidis. Avian Pathol. 1991;20:145–153. - PubMed
-
- Barrow P.A. Rev. Sci. Tech. Vol. 19. 2000. The paratyphoid salmonellae; pp. 351–375. - PubMed
-
- Barrow P.A., Lovell M.A., Lovell M.A. Experimental infection of egg-laying hens with Salmonella enteritidis. Avian Pathol. 1991;20:339–352. - PubMed
-
- Bausch C., Peekhaus N., Utz C., Blais T., Murray E., Lowary T., Conway T., Peekhaus N., Utz C., Blais T., Murray E., Lowary T., Conway T., Utz C., Blais T., Murray E., Lowary T., Conway T., Blais T., Murray E., Lowary T., Conway T., Murray E., Lowary T., Conway T., Lowary T., Conway T., Conway T. Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for L-idonic acid catabolism in Escherichia coli. J. Bacteriol. 1998;180:3704–3710. - PMC - PubMed
-
- Bell K.S., Sebaihia M., Pritchard L., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Sebaihia M., Pritchard L., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Pritchard L., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Bentley S.D., Churcher L.J., Mungall K., Churcher L.J., Mungall K., Mungall K., et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. 2004;101:11105–11110. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
