The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response
- PMID: 18583960
- PMCID: PMC2453059
- DOI: 10.1038/emboj.2008.126
The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response
Abstract
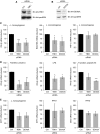
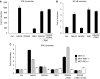
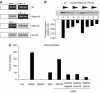
TANK-binding kinase 1 (TBK1) is of central importance for the induction of type-I interferon (IFN) in response to pathogens. We identified the DEAD-box helicase DDX3X as an interaction partner of TBK1. TBK1 and DDX3X acted synergistically in their ability to stimulate the IFN promoter, whereas RNAi-mediated reduction of DDX3X expression led to an impairment of IFN production. Chromatin immunoprecipitation indicated that DDX3X is recruited to the IFN promoter upon infection with Listeria monocytogenes, suggesting a transcriptional mechanism of action. DDX3X was found to be a TBK1 substrate in vitro and in vivo. Phosphorylation-deficient mutants of DDX3X failed to synergize with TBK1 in their ability to stimulate the IFN promoter. Overall, our data imply that DDX3X is a critical effector of TBK1 that is necessary for type I IFN induction.
Figures





References
-
- Akira S (2006) TLR signaling. Curr Top Microbiol Immunol 311: 1–16 - PubMed
-
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G et al. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 - PubMed
-
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

