Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation
- PMID: 18584029
- PMCID: PMC2430619
- DOI: 10.1371/journal.pgen.1000106
Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation
Abstract
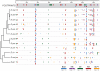
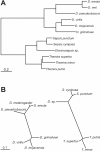
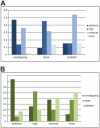
The gene expression pattern specified by an animal regulatory sequence is generally viewed as arising from the particular arrangement of transcription factor binding sites it contains. However, we demonstrate here that regulatory sequences whose binding sites have been almost completely rearranged can still produce identical outputs. We sequenced the even-skipped locus from six species of scavenger flies (Sepsidae) that are highly diverged from the model species Drosophila melanogaster, but share its basic patterns of developmental gene expression. Although there is little sequence similarity between the sepsid eve enhancers and their well-characterized D. melanogaster counterparts, the sepsid and Drosophila enhancers drive nearly identical expression patterns in transgenic D. melanogaster embryos. We conclude that the molecular machinery that connects regulatory sequences to the transcription apparatus is more flexible than previously appreciated. In exploring this diverse collection of sequences to identify the shared features that account for their similar functions, we found a small number of short (20-30 bp) sequences nearly perfectly conserved among the species. These highly conserved sequences are strongly enriched for pairs of overlapping or adjacent binding sites. Together, these observations suggest that the local arrangement of binding sites relative to each other is more important than their overall arrangement into larger units of cis-regulatory function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change.Development. 1998 Mar;125(5):949-58. doi: 10.1242/dev.125.5.949. Development. 1998. PMID: 9449677
-
Bicoid occurrence and Bicoid-dependent hunchback regulation in lower cyclorrhaphan flies.Evol Dev. 2008 Jul-Aug;10(4):413-20. doi: 10.1111/j.1525-142X.2008.00252.x. Evol Dev. 2008. PMID: 18638318
-
Molecular dissection of cis-regulatory modules at the Drosophila bithorax complex reveals critical transcription factor signature motifs.Dev Biol. 2011 Nov 15;359(2):290-302. doi: 10.1016/j.ydbio.2011.07.028. Epub 2011 Jul 28. Dev Biol. 2011. PMID: 21821017 Free PMC article.
-
One thousand and one ways of making functionally similar transcriptional enhancers.Bioessays. 2008 Nov;30(11-12):1052-7. doi: 10.1002/bies.20849. Bioessays. 2008. PMID: 18937349 Review.
-
40 years of homeodomain transcription factors in the Drosophila nervous system.Development. 2024 Jun 1;151(11):dev202910. doi: 10.1242/dev.202910. Epub 2024 May 31. Development. 2024. PMID: 38819456 Free PMC article. Review.
Cited by
-
Intronic cis-regulatory modules mediate tissue-specific and microbial control of angptl4/fiaf transcription.PLoS Genet. 2012;8(3):e1002585. doi: 10.1371/journal.pgen.1002585. Epub 2012 Mar 29. PLoS Genet. 2012. PMID: 22479192 Free PMC article.
-
Conservation of enhancer location in divergent insects.Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14414-9. doi: 10.1073/pnas.0905754106. Epub 2009 Aug 3. Proc Natl Acad Sci U S A. 2009. PMID: 19666595 Free PMC article.
-
Evolutionary mirages: selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers.PLoS Genet. 2010 Jan 22;6(1):e1000829. doi: 10.1371/journal.pgen.1000829. PLoS Genet. 2010. PMID: 20107516 Free PMC article.
-
An alignment-free method to identify candidate orthologous enhancers in multiple Drosophila genomes.Bioinformatics. 2010 Sep 1;26(17):2109-15. doi: 10.1093/bioinformatics/btq358. Epub 2010 Jul 11. Bioinformatics. 2010. PMID: 20624780 Free PMC article.
-
Cis-regulatory evolution integrated the Bric-à-brac transcription factors into a novel fruit fly gene regulatory network.Elife. 2018 Jan 3;7:e32273. doi: 10.7554/eLife.32273. Elife. 2018. PMID: 29297463 Free PMC article.
References
-
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. - PubMed
-
- Costas J, Casares F, Vieira J. Turnover of binding sites for transcription factors involved in early Drosophila development. Gene. 2003;310:215–220. - PubMed
-
- Dermitzakis ET, Bergman CM, Clark AG. Tracing the evolutionary history of Drosophila regulatory regions with models that identify transcription factor binding sites. Mol Biol Evol. 2003;20:703–714. - PubMed
-
- Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol. 2002;19:1114–1121. - PubMed
-
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

