A comparison of BRCT domains involved in nonhomologous end-joining: introducing the solution structure of the BRCT domain of polymerase lambda
- PMID: 18585102
- PMCID: PMC2583787
- DOI: 10.1016/j.dnarep.2008.04.018
A comparison of BRCT domains involved in nonhomologous end-joining: introducing the solution structure of the BRCT domain of polymerase lambda
Abstract
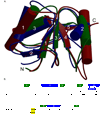
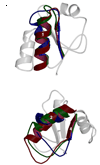
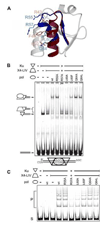
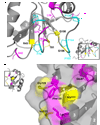
Three of the four family X polymerases, DNA polymerase lambda, DNA polymerase mu, and TdT, have been associated with repair of double-strand DNA breaks by nonhomologous end-joining. Their involvement in this DNA repair process requires an N-terminal BRCT domain that mediates interaction with other protein factors required for recognition and binding of broken DNA ends. Here we present the NMR solution structure of the BRCT domain of DNA polymerase lambda, completing the structural portrait for this family of enzymes. Analysis of the overall fold of the polymerase lambda BRCT domain reveals structural similarity to the BRCT domains of polymerase mu and TdT, yet highlights some key sequence and structural differences that may account for important differences in the biological activities of these enzymes and their roles in nonhomologous end-joining. Mutagenesis studies indicate that the conserved Arg57 residue of Pol lambda plays a more critical role for binding to the XRCC4-Ligase IV complex than its structural homolog in Pol mu, Arg43. In contrast, the hydrophobic Leu60 residue of Pol lambda contributes less significantly to binding than the structurally homologous Phe46 residue of Pol mu. A third leucine residue involved in the binding and activity of Pol mu, is nonconservatively replaced by a glutamine in Pol lambda (Gln64) and, based on binding and activity data, is apparently unimportant for Pol lambda interactions with the NHEJ complex. In conclusion, both the structure of the Pol lambda BRCT domain and its mode of interaction with the other components of the NHEJ complex significantly differ from the two previously studied homologs, Pol mu and TdT.
Figures



References
-
- Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280:18469–18475. - PubMed
-
- Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. - PubMed
-
- Fan W, Wu X. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem Biophys Res Commun. 2004;323:1328–1333. - PubMed
-
- Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

