Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences
- PMID: 18585359
- PMCID: PMC2531161
- DOI: 10.1016/j.cell.2008.05.024
Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences
Abstract
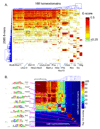
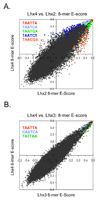
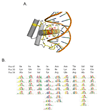
Most homeodomains are unique within a genome, yet many are highly conserved across vast evolutionary distances, implying strong selection on their precise DNA-binding specificities. We determined the binding preferences of the majority (168) of mouse homeodomains to all possible 8-base sequences, revealing rich and complex patterns of sequence specificity and showing that there are at least 65 distinct homeodomain DNA-binding activities. We developed a computational system that successfully predicts binding sites for homeodomain proteins as distant from mouse as Drosophila and C. elegans, and we infer full 8-mer binding profiles for the majority of known animal homeodomains. Our results provide an unprecedented level of resolution in the analysis of this simple domain structure and suggest that variation in sequence recognition may be a factor in its functional diversity and evolutionary success.
Figures




Comment in
-
A lexicon for homeodomain-DNA recognition.Cell. 2008 Jun 27;133(7):1133-5. doi: 10.1016/j.cell.2008.06.008. Cell. 2008. PMID: 18585344
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

