Prediction of drug-target interaction networks from the integration of chemical and genomic spaces
- PMID: 18586719
- PMCID: PMC2718640
- DOI: 10.1093/bioinformatics/btn162
Prediction of drug-target interaction networks from the integration of chemical and genomic spaces
Abstract
Motivation: The identification of interactions between drugs and target proteins is a key area in genomic drug discovery. Therefore, there is a strong incentive to develop new methods capable of detecting these potential drug-target interactions efficiently.
Results: In this article, we characterize four classes of drug-target interaction networks in humans involving enzymes, ion channels, G-protein-coupled receptors (GPCRs) and nuclear receptors, and reveal significant correlations between drug structure similarity, target sequence similarity and the drug-target interaction network topology. We then develop new statistical methods to predict unknown drug-target interaction networks from chemical structure and genomic sequence information simultaneously on a large scale. The originality of the proposed method lies in the formalization of the drug-target interaction inference as a supervised learning problem for a bipartite graph, the lack of need for 3D structure information of the target proteins, and in the integration of chemical and genomic spaces into a unified space that we call 'pharmacological space'. In the results, we demonstrate the usefulness of our proposed method for the prediction of the four classes of drug-target interaction networks. Our comprehensively predicted drug-target interaction networks enable us to suggest many potential drug-target interactions and to increase research productivity toward genomic drug discovery.
Availability: Softwares are available upon request.
Supplementary information: Datasets and all prediction results are available at http://web.kuicr.kyoto-u.ac.jp/supp/yoshi/drugtarget/.
Figures

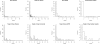
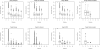
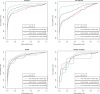

References
-
- Cheng AC, et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007;25:71–75. - PubMed
-
- Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. - PubMed
-
- Gribskov M, Robinson NL. Use of receiver operating characteristic (roc) analysis to evaluate sequence matching. Comput. Chem. 1996;20:25–33. - PubMed
-
- Haggarty SJ, et al. Multidimensional chemical genetic analysis of diversity-oriented synthesis-derived deacetylase inhibitors using cell-based assays. Chem. Biol. 2003;10:383–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

