The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus
- PMID: 18586821
- PMCID: PMC2490736
- DOI: 10.1093/nar/gkn383
The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus
Abstract
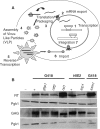
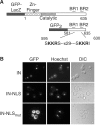
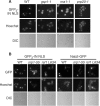
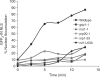
Like its retroviral relatives, the long terminal repeat retrotransposon Ty1 in the yeast Saccharomyces cerevisiae must traverse a permanently intact nuclear membrane for successful transposition and replication. For retrotransposition to occur, at least a subset of Ty1 proteins, including the Ty1 integrase, must enter the nucleus. Nuclear localization of integrase is dependent upon a C-terminal nuclear targeting sequence. However, the nuclear import machinery that recognizes this nuclear targeting signal has not been defined. We investigated the mechanism by which Ty1 integrase gains access to nuclear DNA as a model for how other retroelements, including retroviruses like HIV, may utilize cellular nuclear transport machinery to import their essential nuclear proteins. We show that Ty1 retrotransposition is significantly impaired in yeast mutants that alter the classical nuclear protein import pathway, including the Ran-GTPase, and the dimeric import receptor, importin-alpha/beta. Although Ty1 proteins are made and processed in these mutant cells, our studies reveal that an integrase reporter is not properly targeted to the nucleus in cells carrying mutations in the classical nuclear import machinery. Furthermore, we demonstrate that integrase coimmunoprecipitates with the importin-alpha transport receptor and directly binds to importin-alpha. Taken together, these data suggest Ty1 integrase can employ the classical nuclear protein transport machinery to enter the nucleus.
Figures

References
-
- Greene WC, Peterlin BM. Charting HIV's remarkable voyage through the cell: Basic science as a passport to the future therapy. Nat. Med. 2002;8:673–680. - PubMed
-
- Rijck JD, Vandekerckhove L, Christ F, Debyser Z. Lentiviral nuclear import: a complex interplay between virus and host. BioEssays. 2007;29:441–451. - PubMed
-
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

