Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes
- PMID: 18586827
- PMCID: PMC2490760
- DOI: 10.1093/nar/gkn384
Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes
Abstract
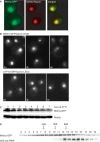
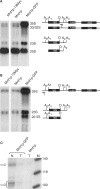
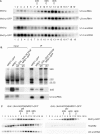
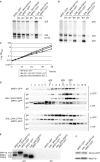
In Saccharomyces cerevisiae, synthesis of the small ribosomal subunit requires assembly of the 35S pre-rRNA into a 90S preribosomal complex. SnoRNAs, including U3 snoRNA, and many trans-acting proteins are required for the ordered assembly and function of the 90S preribosomal complex. Here, we show that the conserved protein Mrd1p binds to the pre-rRNA early during transcription and is required for compaction of the pre-18S rRNA into SSU processome particles. We have exploited the fact that an Mrd1p-GFP fusion protein is incorporated into the 90S preribosomal complex, where it acts as a partial loss-of-function mutation. When associated with the pre-rRNA, Mrd1p-GFP functionally interacts with the essential Pwp2, Mpp10 and U3 snoRNP subcomplexes that are functionally interconnected in the 90S preribosomal complex. The fusion protein can partially support 90S preribosome-mediated cleavages at the A(0)-A(2) sites. At the same time, on a substantial fraction of transcripts, the composition and/or structure of the 90S preribosomal complex is perturbed by the fusion protein in such a way that cleavage of the 35S pre-rRNA is either blocked or shifted to aberrant sites. These results show that Mrd1p is required for establishing productive structures within the 90S preribosomal complex.
Figures



Similar articles
-
Mrd1p is required for release of base-paired U3 snoRNA within the preribosomal complex.Mol Cell Biol. 2009 Nov;29(21):5763-74. doi: 10.1128/MCB.00428-09. Epub 2009 Aug 24. Mol Cell Biol. 2009. PMID: 19704003 Free PMC article.
-
Linker 2 of the eukaryotic pre-ribosomal processing factor Mrd1p is an essential interdomain functionally coupled to upstream RNA Binding Domain 2 (RBD2).PLoS One. 2017 Apr 7;12(4):e0175506. doi: 10.1371/journal.pone.0175506. eCollection 2017. PLoS One. 2017. PMID: 28388671 Free PMC article.
-
Nop6, a component of 90S pre-ribosomal particles, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae.RNA Biol. 2011 Jan-Feb;8(1):112-24. doi: 10.4161/rna.8.1.14143. Epub 2011 Jan 1. RNA Biol. 2011. PMID: 21282979
-
Mpp10p, a new protein component of the U3 snoRNP required for processing of 18S rRNA precursors.Nucleic Acids Symp Ser. 1997;(36):64-7. Nucleic Acids Symp Ser. 1997. PMID: 9478208 Review.
-
Inside the 40S ribosome assembly machinery.Curr Opin Chem Biol. 2011 Oct;15(5):657-63. doi: 10.1016/j.cbpa.2011.07.023. Epub 2011 Aug 20. Curr Opin Chem Biol. 2011. PMID: 21862385 Free PMC article. Review.
Cited by
-
Pwp2 mediates UTP-B assembly via two structurally independent domains.Sci Rep. 2017 Jun 9;7(1):3169. doi: 10.1038/s41598-017-03034-y. Sci Rep. 2017. PMID: 28600509 Free PMC article.
-
Cyclophilin acts as a ribosome biogenesis factor by chaperoning the ribosomal protein (PlRPS15) in filamentous fungi.Nucleic Acids Res. 2021 Dec 2;49(21):12358-12376. doi: 10.1093/nar/gkab1102. Nucleic Acids Res. 2021. PMID: 34792171 Free PMC article.
-
Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit.Mol Cell Biol. 2010 Mar;30(5):1130-44. doi: 10.1128/MCB.00999-09. Epub 2009 Dec 28. Mol Cell Biol. 2010. PMID: 20038530 Free PMC article.
-
The nucleolar protein Nop19p interacts preferentially with Utp25p and Dhr2p and is essential for the production of the 40S ribosomal subunit in Saccharomyces cerevisiae.RNA Biol. 2011 Nov-Dec;8(6):1158-72. doi: 10.4161/rna.8.6.17699. Epub 2011 Nov 1. RNA Biol. 2011. PMID: 21941128 Free PMC article.
-
The small subunit processome in ribosome biogenesis—progress and prospects.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):1-21. doi: 10.1002/wrna.57. Wiley Interdiscip Rev RNA. 2011. PMID: 21318072 Free PMC article. Review.
References
-
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. - PubMed
-
- Udem SR, Warner JR. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J. Mol. Biol. 1972;65:227–242. - PubMed
-
- Trapman J, Retél J, Planta RJ. Ribosomal precursor particles from yeast. Exp. Cell Res. 1975;90:95–104. - PubMed
-
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

