Interplay among side chain sequence, backbone composition, and residue rigidification in polypeptide folding and assembly
- PMID: 18587049
- PMCID: PMC2453709
- DOI: 10.1073/pnas.0801135105
Interplay among side chain sequence, backbone composition, and residue rigidification in polypeptide folding and assembly
Erratum in
- Proc Natl Acad Sci U S A. 2008 Nov 4;105(44):17205
Abstract
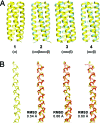
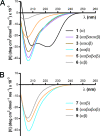
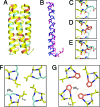
The extent to which polypeptide conformation depends on side-chain composition and sequence has been widely studied, but less is known about the importance of maintaining an alpha-amino acid backbone. Here, we examine a series of peptides with backbones that feature different repeating patterns of alpha- and beta-amino acid residues but an invariant side-chain sequence. In the pure alpha-backbone, this sequence corresponds to the previously studied peptide GCN4-pLI, which forms a very stable four-helix bundle quaternary structure. Physical characterization in solution and crystallographic structure determination show that a variety of alpha/beta-peptide backbones can adopt sequence-encoded quaternary structures similar to that of the alpha prototype. There is a loss in helix bundle stability upon beta-residue incorporation; however, stability of the quaternary structure is not a simple function of beta-residue content. We find that cyclically constrained beta-amino acid residues can stabilize the folds of alpha/beta-peptide GCN4-pLI analogues and restore quaternary structure formation to backbones that are predominantly unfolded in the absence of cyclic residues. Our results show a surprising degree of plasticity in terms of the backbone compositions that can manifest the structural information encoded in a sequence of amino acid side chains. These findings offer a framework for the design of nonnatural oligomers that mimic the structural and functional properties of proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
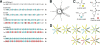
Similar articles
-
Structural consequences of beta-amino acid preorganization in a self-assembling alpha/beta-peptide: fundamental studies of foldameric helix bundles.J Am Chem Soc. 2010 Sep 8;132(35):12378-87. doi: 10.1021/ja103543s. J Am Chem Soc. 2010. PMID: 20718422 Free PMC article.
-
Structural and biological mimicry of protein surface recognition by alpha/beta-peptide foldamers.Proc Natl Acad Sci U S A. 2009 Sep 1;106(35):14751-6. doi: 10.1073/pnas.0902663106. Epub 2009 Aug 17. Proc Natl Acad Sci U S A. 2009. PMID: 19706443 Free PMC article.
-
Foldamers with heterogeneous backbones.Acc Chem Res. 2008 Oct;41(10):1399-408. doi: 10.1021/ar800009n. Epub 2008 Jul 1. Acc Chem Res. 2008. PMID: 18590282 Free PMC article.
-
Gabapentin: a stereochemically constrained gamma amino acid residue in hybrid peptide design.Acc Chem Res. 2009 Oct 20;42(10):1628-39. doi: 10.1021/ar9001153. Acc Chem Res. 2009. PMID: 19572698 Review.
-
The world of beta- and gamma-peptides comprised of homologated proteinogenic amino acids and other components.Chem Biodivers. 2004 Aug;1(8):1111-239. doi: 10.1002/cbdv.200490087. Chem Biodivers. 2004. PMID: 17191902 Review.
Cited by
-
α-Helix mimicry with α/β-peptides.Methods Enzymol. 2013;523:407-29. doi: 10.1016/B978-0-12-394292-0.00019-9. Methods Enzymol. 2013. PMID: 23422441 Free PMC article.
-
The Diverse World of Foldamers: Endless Possibilities of Self-Assembly.Molecules. 2020 Jul 18;25(14):3276. doi: 10.3390/molecules25143276. Molecules. 2020. PMID: 32708440 Free PMC article. Review.
-
Altered signaling at the PTH receptor via modified agonist contacts with the extracellular domain provides a path to prolonged agonism in vivo.Proc Natl Acad Sci U S A. 2022 Nov 29;119(48):e2212736119. doi: 10.1073/pnas.2212736119. Epub 2022 Nov 21. Proc Natl Acad Sci U S A. 2022. PMID: 36409914 Free PMC article.
-
Receptor selectivity from minimal backbone modification of a polypeptide agonist.Proc Natl Acad Sci U S A. 2018 Dec 4;115(49):12383-12388. doi: 10.1073/pnas.1815294115. Epub 2018 Nov 15. Proc Natl Acad Sci U S A. 2018. PMID: 30442659 Free PMC article.
-
Development of Potent, Protease-Resistant Agonists of the Parathyroid Hormone Receptor with Broad β Residue Distribution.J Med Chem. 2017 Nov 9;60(21):8816-8833. doi: 10.1021/acs.jmedchem.7b00876. Epub 2017 Oct 24. J Med Chem. 2017. PMID: 29064243 Free PMC article.
References
-
- Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D. Progress on modeling of protein structures and interactions. Science. 2005;310:638–642. - PubMed
-
- Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr Opin Struct Biol. 2007;17:157–165. - PubMed
-
- Eschenmoser A. Chemical etiology of nucleic acid structure. Science. 1999;284:2118–2124. - PubMed
-
- Cheng RP, Gellman SH, DeGrado WF. β-Peptides: From structure to function. Chem Rev. 2001;101:3219–3232. - PubMed
-
- Seebach D, Beck AK, Bierbaum DJ. The world of β- and γ-peptides comprised of homologated proteinogenic amino acids and other components. Chem Biodivers. 2004;1:1111–1239. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

