High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids
- PMID: 18587050
- PMCID: PMC2453694
- DOI: 10.1073/pnas.0800406105
High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids
Abstract
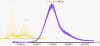
Hepatitis B virus (HBV) is a major human pathogen. In addition to its importance in human health, there is growing interest in adapting HBV and other viruses for drug delivery and other nanotechnological applications. In both contexts, precise biophysical characterization of these large macromolecular particles is fundamental. HBV capsids are unusual in that they exhibit two distinct icosahedral geometries, nominally composed of 90 and 120 dimers with masses of approximately 3 and approximately 4 MDa, respectively. Here, a mass spectrometric approach was used to determine the masses of both capsids to within 0.1%. It follows that both lattices are complete, consisting of exactly 180 and 240 subunits. Nanoindentation experiments by atomic-force microscopy indicate that both capsids have similar stabilities. The data yielded a Young's modulus of approximately 0.4 GPa. This experimental approach, anchored on very precise and accurate mass measurements, appears to hold considerable potential for elucidating the assembly of viruses and other macromolecular particles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
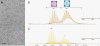

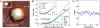
References
-
- Deres K, et al. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299:893–896. - PubMed
-
- Singh P, Gonzalez MJ, Manchester M. Viruses and their uses in nanotechnology. Drug Dev Res. 2006;67:23–41.
-
- Steven AC, et al. Structure, assembly, and antigenicity of hepatitis B virus capsid proteins. Adv Virus Res. 2005;64:125–164. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

