Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis
- PMID: 18587411
- PMCID: PMC2764292
- DOI: 10.1038/nrmicro1928
Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis
Abstract
Viruses of the Flaviviridae family, including hepatitis C, dengue and bovine viral diarrhoea, are responsible for considerable morbidity and mortality worldwide. Recent advances in our understanding of virion assembly have uncovered commonalities among distantly related members of this family. We discuss the emerging hypothesis that physical virion components are not alone in forming the infectious particle, but that non-structural proteins are intimately involved in orchestrating morphogenesis. Pinpointing the roles of Flaviviridae proteins in virion production could reveal new avenues for antiviral therapeutics.
Figures
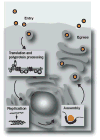
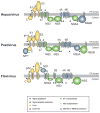
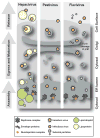
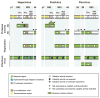

References
-
- Lindenbach BD, Thiel HJ, Rice CM. In: Fields Virology. Knipe DM, Howley PM, editors. Lippincott-Raven Publishers; Philadelphia: 2007. pp. 1101–1152.
-
- Gray EW, Nettleton PF. The ultrastructure of cell cultures infected with border disease and bovine virus diarrhoea viruses. J Gen Virol. 1987;68:2339–2346. - PubMed
-
- Mottola G, Cardinali G, Ceccacci A, Trozzi C, Bartholomew L, Torrisi MR, Pedrazzini E, Bonatti S, Migliaccio G. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology. 2002;293:31–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

