PMC42, a breast progenitor cancer cell line, has normal-like mRNA and microRNA transcriptomes
- PMID: 18588681
- PMCID: PMC2481505
- DOI: 10.1186/bcr2109
PMC42, a breast progenitor cancer cell line, has normal-like mRNA and microRNA transcriptomes
Abstract
Introduction: The use of cultured cell lines as model systems for normal tissue is limited by the molecular alterations accompanying the immortalisation process, including changes in the mRNA and microRNA (miRNA) repertoire. Therefore, identification of cell lines with normal-like expression profiles is of paramount importance in studies of normal gene regulation.
Methods: The mRNA and miRNA expression profiles of several breast cell lines of cancerous or normal origin were measured using printed slide arrays, Luminex bead arrays, and real-time reverse transcription-polymerase chain reaction.
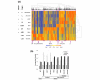
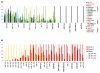
Results: We demonstrate that the mRNA expression profiles of two breast cell lines are similar to that of normal breast tissue: HB4a, immortalised normal breast epithelium, and PMC42, a breast cancer cell line that retains progenitor pluripotency allowing in-culture differentiation to both secretory and myoepithelial fates. In contrast, only PMC42 exhibits a normal-like miRNA expression profile. We identified a group of miRNAs that are highly expressed in normal breast tissue and PMC42 but are lost in all other cancerous and normal-origin breast cell lines and observed a similar loss in immortalised lymphoblastoid cell lines compared with healthy uncultured B cells. Moreover, like tumour suppressor genes, these miRNAs are lost in a variety of tumours. We show that the mechanism leading to the loss of these miRNAs in breast cancer cell lines has genomic, transcriptional, and post-transcriptional components.
Conclusion: We propose that, despite its neoplastic origin, PMC42 is an excellent molecular model for normal breast epithelium, providing a unique tool to study breast differentiation and the function of key miRNAs that are typically lost in cancer.
Figures
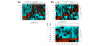


References
-
- Bissell MJ. The differentiated state of normal and malignant cells or how to define a 'normal' cell in culture. Int Rev Cytol. 1981;70:27–100. - PubMed
-
- Whitehead RH, Bertoncello I, Webber LM, Pedersen JS. A new human breast carcinoma cell line (PMC42) with stem cell characteristics. I. Morphologic characterization. J Natl Cancer Inst. 1983;70:649–661. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

