Comparative genomic profiling of Dutch clinical Bordetella pertussis isolates using DNA microarrays: identification of genes absent from epidemic strains
- PMID: 18590534
- PMCID: PMC2481270
- DOI: 10.1186/1471-2164-9-311
Comparative genomic profiling of Dutch clinical Bordetella pertussis isolates using DNA microarrays: identification of genes absent from epidemic strains
Erratum in
- BMC Genomics. 2010;11:196
Abstract
Background: Whooping cough caused by Bordetella pertussis in humans, is re-emerging in many countries despite vaccination. Several studies have shown that significant shifts have occurred in the B. pertussis population resulting in antigenic divergence between vaccine strains and circulating strains and suggesting pathogen adaptation. In the Netherlands, the resurgence of pertussis is associated with the rise of B. pertussis strains with an altered promoter region for pertussis toxin (ptxP3).
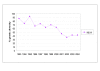
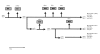
Results: We used Multi-Locus Sequence Typing (MLST), Multiple-Locus Variable Number of Tandem Repeat Analysis (MLVA) and microarray-based comparative genomic hybridization (CGH) to characterize the ptxP3 strains associated with the Dutch epidemic. For CGH analysis, we developed an oligonucleotide (70-mers) microarray consisting of 3,581 oligonucleotides representing 94% of the gene repertoire of the B. pertussis strain Tohama I. Nine different MLST profiles and 38 different MLVA types were found in the period 1993 to 2004. Forty-three Dutch clinical isolates were analyzed with CGH, 98 genes were found to be absent in at least one of the B. pertussis strains tested, these genes were clustered in 8 distinct regions of difference.
Conclusion: The presented MLST, MLVA and CGH-analysis identified distinctive characteristics of ptxP3 B. pertussis strains -the most prominent of which was a genomic deletion removing about 23,000 bp. We propose a model for the emergence of ptxP3 strains.
Figures




Similar articles
-
Analysis of Swedish Bordetella pertussis isolates with three typing methods: characterization of an epidemic lineage.J Microbiol Methods. 2009 Sep;78(3):297-301. doi: 10.1016/j.mimet.2009.06.019. Epub 2009 Jul 3. J Microbiol Methods. 2009. PMID: 19577594
-
Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s.J Bacteriol. 2004 Aug;186(16):5496-505. doi: 10.1128/JB.186.16.5496-5505.2004. J Bacteriol. 2004. PMID: 15292152 Free PMC article.
-
Differences in the genomic content of Bordetella pertussis isolates before and after introduction of pertussis vaccines in four European countries.Infect Genet Evol. 2011 Dec;11(8):2034-42. doi: 10.1016/j.meegid.2011.09.012. Epub 2011 Sep 21. Infect Genet Evol. 2011. PMID: 21964035
-
Epidemiology of whooping cough & typing of Bordetella pertussis.Future Microbiol. 2013 Nov;8(11):1391-403. doi: 10.2217/fmb.13.111. Future Microbiol. 2013. PMID: 24199799 Review.
-
Molecular Epidemiology of Bordetella pertussis.Adv Exp Med Biol. 2019;1183:19-33. doi: 10.1007/5584_2019_402. Adv Exp Med Biol. 2019. PMID: 31342459 Review.
Cited by
-
Comparative genomics of prevaccination and modern Bordetella pertussis strains.BMC Genomics. 2010 Nov 11;11:627. doi: 10.1186/1471-2164-11-627. BMC Genomics. 2010. PMID: 21070624 Free PMC article.
-
Strategies and new developments to control pertussis, an actual health problem.Pathog Dis. 2015 Nov;73(8):ftv059. doi: 10.1093/femspd/ftv059. Epub 2015 Aug 9. Pathog Dis. 2015. PMID: 26260328 Free PMC article. Review.
-
Bordetella pertussis strains with increased toxin production associated with pertussis resurgence.Emerg Infect Dis. 2009 Aug;15(8):1206-13. doi: 10.3201/eid1508.081511. Emerg Infect Dis. 2009. PMID: 19751581 Free PMC article.
-
Bordetella pertussis Isolates from Argentinean Whooping Cough Patients Display Enhanced Biofilm Formation Capacity Compared to Tohama I Reference Strain.Front Microbiol. 2015 Dec 8;6:1352. doi: 10.3389/fmicb.2015.01352. eCollection 2015. Front Microbiol. 2015. PMID: 26696973 Free PMC article.
-
Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: elucidation of factors involved in the increased fitness of epidemic strains.PLoS One. 2013 Jun 11;8(6):e66150. doi: 10.1371/journal.pone.0066150. Print 2013. PLoS One. 2013. PMID: 23776625 Free PMC article.
References
-
- van der Zee A, Groenendijk H, Peeters M, Mooi FR. The differentiation of Bordetella parapertussis and Bordetella bronchiseptica from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. Int J Syst Bacteriol. 1996;46:640–647. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

