Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus
- PMID: 18590559
- PMCID: PMC2491623
- DOI: 10.1186/1471-2180-8-110
Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus
Abstract
Background: Vibrio parahaemolyticus is abundant in the aquatic environment particularly in warmer waters and is the leading cause of seafood borne gastroenteritis worldwide. Prior to 1995, numerous V. parahaemolyticus serogroups were associated with disease, however, in that year an O3:K6 serogroup emerged in Southeast Asia causing large outbreaks and rapid hospitalizations. This new highly virulent strain is now globally disseminated.
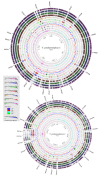
Results: We performed a four-way BLAST analysis on the genome sequence of V. parahaemolyticus RIMD2210633, an O3:K6 isolate from Japan recovered in 1996, versus the genomes of four published Vibrio species and constructed genome BLAST atlases. We identified 24 regions, gaps in the genome atlas, of greater than 10 kb that were unique to RIMD2210633. These 24 regions included an integron, f237 phage, 2 type III secretion systems (T3SS), a type VI secretion system (T6SS) and 7 Vibrio parahaemolyticus genomic islands (VPaI-1 to VPaI-7). Comparative genomic analysis of our fifth genome, V. parahaemolyticus AQ3810, an O3:K6 isolate recovered in 1983, identified four regions unique to each V. parahaemolyticus strain. Interestingly, AQ3810 did not encode 8 of the 24 regions unique to RMID, including a T6SS, which suggests an additional virulence mechanism in RIMD2210633. The distribution of only the VPaI regions was highly variable among a collection of 42 isolates and phylogenetic analysis of these isolates show that these regions are confined to a pathogenic clade.
Conclusion: Our data show that there is considerable genomic flux in this species and that the new highly virulent clone arose from an O3:K6 isolate that acquired at least seven novel regions, which included both a T3SS and a T6SS.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

