Pmp-like proteins Pls1 and Pls2 are secreted into the lumen of the Chlamydia trachomatis inclusion
- PMID: 18591235
- PMCID: PMC2519427
- DOI: 10.1128/IAI.00632-08
Pmp-like proteins Pls1 and Pls2 are secreted into the lumen of the Chlamydia trachomatis inclusion
Abstract
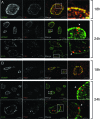
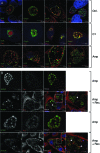
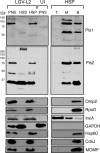
The obligate intracellular pathogen Chlamydia trachomatis secretes effector proteins across the membrane of the pathogen-containing vacuole (inclusion) to modulate host cellular functions. In an immunological screen for secreted chlamydial proteins, we identified CT049 and CT050 as potential inclusion membrane-associated proteins. These acidic, nonglobular proteins are paralogously related to the passenger domain of the polymorphic membrane protein PmpC and, like other Pmp proteins, are highly polymorphic among C. trachomatis ocular and urogenital strains. We generated antibodies to these Pmp-like secreted (Pls) proteins and determined by immunofluorescence microscopy that Pls1 (CT049) and Pls2 (CT050) localized to globular structures within the inclusion lumen and at the inclusion membrane. Fractionation of membranes and cytoplasmic components from infected cells by differential and density gradient centrifugation further indicated that Pls1 and Pls2 associated with membranes distinct from the bulk of bacterial and inclusion membranes. The accumulation of Pls1 and, to a lesser extent, Pls2 in the inclusion lumen was insensitive to the type III secretion inhibitor C1, suggesting that this translocation system is not essential for Pls protein secretion. In contrast, Pls secretion and stability were sensitive to low levels of beta-lactam antibiotics, suggesting that a functional cell wall is required for Pls secretion from the bacterial cell. Finally, we tested the requirement for these proteins in Chlamydia infection by microinjecting anti-Pls1 and anti-Pls2 antibodies into infected cells. Coinjection of anti-Pls1 and -Pls2 antibodies partially inhibited expansion of the inclusion. Because Pls proteins lack classical sec-dependent secretion signals, we propose that Pls proteins are secreted into the inclusion lumen by a novel mechanism to regulate events important for chlamydial replication and inclusion expansion.
Figures




Similar articles
-
The Human Centrosomal Protein CCDC146 Binds Chlamydia trachomatis Inclusion Membrane Protein CT288 and Is Recruited to the Periphery of the Chlamydia-Containing Vacuole.Front Cell Infect Microbiol. 2018 Jul 26;8:254. doi: 10.3389/fcimb.2018.00254. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30094225 Free PMC article.
-
A meta-analysis of affinity purification-mass spectrometry experimental systems used to identify eukaryotic and chlamydial proteins at the Chlamydia trachomatis inclusion membrane.J Proteomics. 2020 Feb 10;212:103595. doi: 10.1016/j.jprot.2019.103595. Epub 2019 Nov 21. J Proteomics. 2020. PMID: 31760040 Free PMC article.
-
Chlamydia trachomatis Inc Ct226 is vital for FLI1 and LRRF1 recruitment to the chlamydial inclusion.mSphere. 2024 Nov 21;9(11):e0047324. doi: 10.1128/msphere.00473-24. Epub 2024 Oct 15. mSphere. 2024. PMID: 39404459 Free PMC article.
-
Got mutants? How advances in chlamydial genetics have furthered the study of effector proteins.Pathog Dis. 2021 Feb 4;79(2):ftaa078. doi: 10.1093/femspd/ftaa078. Pathog Dis. 2021. PMID: 33512479 Free PMC article. Review.
-
Chlamydial polymorphic membrane proteins: regulation, function and potential vaccine candidates.Virulence. 2016;7(1):11-22. doi: 10.1080/21505594.2015.1111509. Epub 2015 Nov 18. Virulence. 2016. PMID: 26580416 Free PMC article. Review.
Cited by
-
Plasmid-cured Chlamydia caviae activates TLR2-dependent signaling and retains virulence in the guinea pig model of genital tract infection.PLoS One. 2012;7(1):e30747. doi: 10.1371/journal.pone.0030747. Epub 2012 Jan 24. PLoS One. 2012. PMID: 22292031 Free PMC article.
-
Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems.PLoS Pathog. 2009 Apr;5(4):e1000375. doi: 10.1371/journal.ppat.1000375. Epub 2009 Apr 24. PLoS Pathog. 2009. PMID: 19390620 Free PMC article.
-
Killing me softly: chlamydial use of proteolysis for evading host defenses.Trends Microbiol. 2009 Oct;17(10):467-74. doi: 10.1016/j.tim.2009.07.007. Epub 2009 Sep 16. Trends Microbiol. 2009. PMID: 19765998 Free PMC article. Review.
-
Genomic Analysis of MSM Rectal Chlamydia trachomatis Isolates Identifies Predicted Tissue-Tropic Lineages Generated by Intraspecies Lateral Gene Transfer-Mediated Evolution.Infect Immun. 2022 Nov 17;90(11):e0026522. doi: 10.1128/iai.00265-22. Epub 2022 Oct 10. Infect Immun. 2022. PMID: 36214558 Free PMC article.
-
A Chlamydial Plasmid-Dependent Secretion System for the Delivery of Virulence Factors to the Host Cytosol.mBio. 2021 Jun 29;12(3):e0117921. doi: 10.1128/mBio.01179-21. Epub 2021 Jun 8. mBio. 2021. PMID: 34101486 Free PMC article.
References
-
- Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 235-47. - PubMed
-
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

