Structure-function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTX toxin
- PMID: 18591243
- PMCID: PMC3259750
- DOI: 10.1074/jbc.M803334200
Structure-function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTX toxin
Abstract
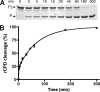
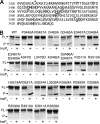
Vibrio cholerae secretes a large virulence-associated multifunctional autoprocessing RTX toxin (MARTX(Vc)). Autoprocessing of this toxin by an embedded cysteine protease domain (CPD) is essential for this toxin to induce actin depolymerization in a broad range of cell types. A homologous CPD is also present in the large clostridial toxin TcdB and recent studies showed that inositol hexakisphosphate (Ins(1,2,3,4,5,6)P(6) or InsP(6)) stimulated the autoprocessing of TcdB dependent upon the CPD (Egerer, M., Giesemann, T., Jank, T., Satchell, K. J., and Aktories, K. (2007) J. Biol. Chem. 282, 25314-25321). In this work, the autoprocessing activity of the CPD within MARTX(Vc) is similarly found to be inducible by InsP(6). The CPD is shown to bind InsP(6) (K(d), 0.6 microm), and InsP(6) is shown to stimulate intramolecular autoprocessing at both physiological concentrations and as low as 0.01 microm. Processed CPD did not bind InsP(6) indicating that, subsequent to cleavage, the activated CPD may shift to an inactive conformation. To further pursue the mechanism of autoprocessing, conserved residues among 24 identified CPDs were mutagenized. In addition to cysteine and histidine residues that form the catalytic site, 2 lysine residues essential for InsP(6) binding and 5 lysine and arginine residues resulting in loss of activity at low InsP(6) concentrations were identified. Overall, our data support a model in which basic residues located across the CPD structure form an InsP(6) binding pocket and that the binding of InsP(6) stimulates processing by altering the CPD to an activated conformation. After processing, InsP(6) is shown to be recycled, while the cleaved CPD becomes incapable of further binding of InsP(6).
Figures





Similar articles
-
Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites.J Biol Chem. 2009 Sep 25;284(39):26557-68. doi: 10.1074/jbc.M109.025510. Epub 2009 Jul 20. J Biol Chem. 2009. PMID: 19620709 Free PMC article.
-
Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain.Science. 2008 Oct 10;322(5899):265-8. doi: 10.1126/science.1162403. Science. 2008. PMID: 18845756 Free PMC article.
-
Autoproteolytic activation of bacterial toxins.Toxins (Basel). 2010 May;2(5):963-77. doi: 10.3390/toxins2050963. Epub 2010 May 6. Toxins (Basel). 2010. PMID: 22069620 Free PMC article. Review.
-
Inositol hexakisphosphate-induced autoprocessing of large bacterial protein toxins.PLoS Pathog. 2010 Jul 8;6(7):e1000942. doi: 10.1371/journal.ppat.1000942. PLoS Pathog. 2010. PMID: 20628577 Free PMC article. Review.
-
Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain.EMBO J. 2007 May 16;26(10):2552-61. doi: 10.1038/sj.emboj.7601700. Epub 2007 Apr 26. EMBO J. 2007. PMID: 17464284 Free PMC article.
Cited by
-
Allosteric regulation of protease activity by small molecules.Mol Biosyst. 2010 Aug;6(8):1431-43. doi: 10.1039/c003913f. Epub 2010 Jun 10. Mol Biosyst. 2010. PMID: 20539873 Free PMC article. Review.
-
Inositol hexakisphosphate-dependent processing of Clostridium sordellii lethal toxin and Clostridium novyi alpha-toxin.J Biol Chem. 2011 Apr 29;286(17):14779-86. doi: 10.1074/jbc.M110.200691. Epub 2011 Mar 8. J Biol Chem. 2011. PMID: 21385871 Free PMC article.
-
Vibrio type III effector VPA1380 is related to the cysteine protease domain of large bacterial toxins.PLoS One. 2014 Aug 6;9(8):e104387. doi: 10.1371/journal.pone.0104387. eCollection 2014. PLoS One. 2014. PMID: 25099122 Free PMC article.
-
Cytotoxicity of the Vibrio vulnificus MARTX toxin effector DUF5 is linked to the C2A subdomain.Proteins. 2014 Oct;82(10):2643-56. doi: 10.1002/prot.24628. Epub 2014 Jun 26. Proteins. 2014. PMID: 24935440 Free PMC article.
-
Host S-nitrosylation inhibits clostridial small molecule-activated glucosylating toxins.Nat Med. 2011 Aug 21;17(9):1136-41. doi: 10.1038/nm.2405. Nat Med. 2011. PMID: 21857653 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

