Convergent evolutionary analysis identifies significant mutations in drug resistance targets of Mycobacterium tuberculosis
- PMID: 18591265
- PMCID: PMC2533473
- DOI: 10.1128/AAC.00309-08
Convergent evolutionary analysis identifies significant mutations in drug resistance targets of Mycobacterium tuberculosis
Abstract
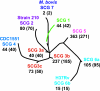
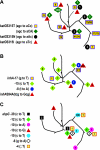
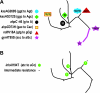
Mycobacterium tuberculosis adapts to the environment by selecting for advantageous single-nucleotide polymorphisms (SNPs). We studied whether advantageous SNPs could be distinguished from neutral mutations within genes associated with drug resistance. A total of 1,003 clinical isolates of M. tuberculosis were related phylogenetically and tested for the distribution of SNPs in putative drug resistance genes. Drug resistance-associated versus non-drug-resistance-associated SNPs in putative drug resistance genes were compared for associations with single versus multiple-branch outcomes using the chi-square and Fisher exact tests. All 286 (100%) isolates containing isoniazid (INH) resistance-associated SNPs had multibranch distributions, suggestive of multiple ancestry and convergent evolution. In contrast, all 327 (100%) isolates containing non-drug-resistance-associated SNPs were monophyletic and thus showed no evidence of convergent evolution (P < 0.001). Convergence testing was then applied to SNPs at position 481 of the iniA (Rv0342) gene and position 306 of the embB gene, both potential drug resistance targets for INH and/or ethambutol. Mutant embB306 alleles showed multibranch distributions, suggestive of convergent evolution; however, all 44 iniA(H481Q) mutations were monophyletic. In conclusion, this study validates convergence analysis as a tool for identifying mutations that cause INH resistance and explores mutations in other genes. Our results suggest that embB306 mutations are likely to confer drug resistance, while iniA(H481Q) mutations are not. This approach may be applied on a genome-wide scale to identify SNPs that impact antibiotic resistance and other types of biological fitness.
Figures

References
-
- Alland, D., D. W. Lacher, M. H. Hazbon, A. S. Motiwala, W. Qi, R. D. Fleischmann, and T. S. Whittam. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 45:39-46. - PMC - PubMed
-
- Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbón, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. - PMC - PubMed
-
- Azhikina, T., N. Gvozdevsky, A. Botvinnik, A. Fushan, I. Shemyakin, V. Stepanshina, M. Lipin, C. Barry III, and E. Sverdlov. 2006. A genome-wide sequence-independent comparative analysis of insertion-deletion polymorphisms in multiple Mycobacterium tuberculosis strains. Res. Microbiol. 157:282-290. - PubMed
-
- Baker, L. V., T. J. Brown, O. Maxwell, A. L. Gibson, Z. Fang, M. D. Yates, and F. A. Drobniewski. 2005. Molecular analysis of isoniazid-resistant Mycobacterium tuberculosis isolates from England and Wales reveals the phylogenetic significance of the ahpC −46A polymorphism. Antimicrob. Agents Chemother. 49:1455-1464. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

