African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping
- PMID: 18593304
- PMCID: PMC2953791
- DOI: 10.1146/annurev.genom.9.081307.164258
African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping
Abstract
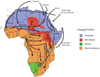
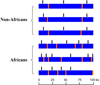
Comparative studies of ethnically diverse human populations, particularly in Africa, are important for reconstructing human evolutionary history and for understanding the genetic basis of phenotypic adaptation and complex disease. African populations are characterized by greater levels of genetic diversity, extensive population substructure, and less linkage disequilibrium (LD) among loci compared to non-African populations. Africans also possess a number of genetic adaptations that have evolved in response to diverse climates and diets, as well as exposure to infectious disease. This review summarizes patterns and the evolutionary origins of genetic diversity present in African populations, as well as their implications for the mapping of complex traits, including disease susceptibility.
Figures

References
-
- Abecasis GR, Ghosh D, Nichols TE. Linkage disequilibrium: ancient history drives the new genetics. Hum. Hered. 2005;59:118–124. - PubMed
-
- Abubakari AR, Lauder W, Agyemang C, Jones M, Kirk A, Bhopal RS. Prevalence and time trends in obesity among adult West African populations: a meta-analysis. Obes. Rev. 2008 doi:10.1111/j.1467-789X.2007.00462.x. - PubMed
-
- Abu-Raddad LJ, Patnaik P, Kublin JG. Dual infection with HIV and malaria fuels the spread of both diseases in sub-Saharan Africa. Science. 2006;314:1603–1606. - PubMed
-
- Adeyemo A, Luke A, Cooper R, Wu X, Tayo B, et al. A genome-wide scan for body mass index among Nigerian families. Obes. Res. 2003;11:266–273. - PubMed
-
-
Deleted in proof.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

