The complete mitochondrial genome of the Antarctic springtail Cryptopygus antarcticus (Hexapoda: Collembola)
- PMID: 18593463
- PMCID: PMC2483729
- DOI: 10.1186/1471-2164-9-315
The complete mitochondrial genome of the Antarctic springtail Cryptopygus antarcticus (Hexapoda: Collembola)
Abstract
Background: Mitogenomics data, i.e. complete mitochondrial genome sequences, are popular molecular markers used for phylogenetic, phylogeographic and ecological studies in different animal lineages. Their comparative analysis has been used to shed light on the evolutionary history of given taxa and on the molecular processes that regulate the evolution of the mitochondrial genome. A considerable literature is available in the fields of invertebrate biochemical and ecophysiological adaptation to extreme environmental conditions, exemplified by those of the Antarctic. Nevertheless, limited molecular data are available from terrestrial Antarctic species, and this study represents the first attempt towards the description of a mitochondrial genome from one of the most widespread and common collembolan species of Antarctica.
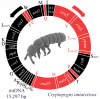
Results: In this study we describe the mitochondrial genome of the Antarctic collembolan Cryptopygus antarcticus Willem, 1901. The genome contains the standard set of 37 genes usually present in animal mtDNAs and a large non-coding fragment putatively corresponding to the region (A+T-rich) responsible for the control of replication and transcription. All genes are arranged in the gene order typical of Pancrustacea. Three additional short non-coding regions are present at gene junctions. Two of these are located in positions of abrupt shift of the coding polarity of genes oriented on opposite strands suggesting a role in the attenuation of the polycistronic mRNA transcription(s). In addition, remnants of an additional copy of trnL(uag) are present between trnS(uga) and nad1. Nucleotide composition is biased towards a high A% and T% (A+T = 70.9%), as typically found in hexapod mtDNAs. There is also a significant strand asymmetry, with the J-strand being more abundant in A and C. Within the A+T-rich region, some short sequence fragments appear to be similar (in position and primary sequence) to those involved in the origin of the N-strand replication of the Drosophila mtDNA.
Conclusion: The mitochondrial genome of C. antarcticus shares several features with other pancrustacean genomes, although the presence of unusual non-coding regions is also suggestive of molecular rearrangements that probably occurred before the differentiation of major collembolan families. Closer examination of gene boundaries also confirms previous observations on the presence of unusual start and stop codons, and suggests a role for tRNA secondary structures as potential cleavage signals involved in the maturation of the primary transcript. Sequences potentially involved in the regulation of replication/transcription are present both in the A+T-rich region and in other areas of the genome. Their position is similar to that observed in a limited number of insect species, suggesting unique replication/transcription mechanisms for basal and derived hexapod lineages. This initial description and characterization of the mitochondrial genome of C. antarcticus will constitute the essential foundation prerequisite for investigations of the evolutionary history of one of the most speciose collembolan genera present in Antarctica and other localities of the Southern Hemisphere.
Figures



Similar articles
-
The complete mitochondrial DNA sequence of the basal hexapod Tetrodontophora bielanensis: evidence for heteroplasmy and tRNA translocations.Mol Biol Evol. 2001 Jul;18(7):1293-304. doi: 10.1093/oxfordjournals.molbev.a003914. Mol Biol Evol. 2001. PMID: 11420368
-
The mitochondrial genome of the ascalaphid owlfly Libelloides macaronius and comparative evolutionary mitochondriomics of neuropterid insects.BMC Genomics. 2011 May 10;12:221. doi: 10.1186/1471-2164-12-221. BMC Genomics. 2011. PMID: 21569260 Free PMC article.
-
The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae).BMC Genomics. 2008 Jul 15;9:331. doi: 10.1186/1471-2164-9-331. BMC Genomics. 2008. PMID: 18627592 Free PMC article.
-
The Drosophila mitochondrial genome.Oxf Surv Eukaryot Genes. 1984;1:1-35. Oxf Surv Eukaryot Genes. 1984. PMID: 6400770 Review.
-
Mitochondrial-derived peptides: Antidiabetic functions and evolutionary perspectives.Peptides. 2024 Feb;172:171147. doi: 10.1016/j.peptides.2023.171147. Epub 2023 Dec 29. Peptides. 2024. PMID: 38160808 Free PMC article. Review.
Cited by
-
The complete mitochondrial genome of the citrus red mite Panonychus citri (Acari: Tetranychidae): high genome rearrangement and extremely truncated tRNAs.BMC Genomics. 2010 Oct 23;11:597. doi: 10.1186/1471-2164-11-597. BMC Genomics. 2010. PMID: 20969792 Free PMC article.
-
Complete Mitochondrial Genome of Suwallia teleckojensis (Plecoptera: Chloroperlidae) and Implications for the Higher Phylogeny of Stoneflies.Int J Mol Sci. 2018 Feb 28;19(3):680. doi: 10.3390/ijms19030680. Int J Mol Sci. 2018. PMID: 29495588 Free PMC article.
-
Analysis of the genome of the New Zealand giant collembolan (Holacanthella duospinosa) sheds light on hexapod evolution.BMC Genomics. 2017 Oct 17;18(1):795. doi: 10.1186/s12864-017-4197-1. BMC Genomics. 2017. PMID: 29041914 Free PMC article.
-
Next-generation sequencing, phylogenetic signal and comparative mitogenomic analyses in Metacrangonyctidae (Amphipoda: Crustacea).BMC Genomics. 2014 Jul 6;15(1):566. doi: 10.1186/1471-2164-15-566. BMC Genomics. 2014. PMID: 24997985 Free PMC article.
-
Novel insights into mitochondrial gene rearrangement in thrips (Insecta: Thysanoptera) from the grass thrips, Anaphothrips obscurus.Sci Rep. 2017 Jun 27;7(1):4284. doi: 10.1038/s41598-017-04617-5. Sci Rep. 2017. PMID: 28655921 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

