Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi
- PMID: 18593478
- PMCID: PMC2474616
- DOI: 10.1186/1471-2148-8-190
Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi
Abstract
Background: The evolution of multicellularity is accompanied by the occurrence of differentiated tissues, of organismal developmental programs, and of mechanisms keeping the balance between proliferation and differentiation. Initially, the SET-domain proteins were associated exclusively with regulation of developmental genes in metazoa. However, finding of SET-domain genes in the unicellular yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe suggested that SET-domain proteins regulate a much broader variety of biological programs. Intuitively, it is expected that the numbers, types, and biochemical specificity of SET-domain proteins of multicellular versus unicellular forms would reflect the differences in their biology. However, comparisons across the unicellular and multicellular domains of life are complicated by the lack of knowledge of the ancestral SET-domain genes. Even within the crown group, different biological systems might use the epigenetic 'code' differently, adapting it to organism-specific needs. Simplifying the model, we undertook a systematic phylogenetic analysis of one monophyletic fungal group (Ascomycetes) containing unicellular yeasts, Saccharomycotina (hemiascomycetes), and a filamentous fungal group, Pezizomycotina (euascomycetes).
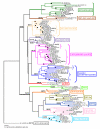
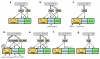
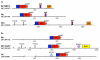
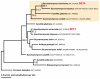
Results: Systematic analysis of the SET-domain genes across an entire eukaryotic phylum has outlined clear distinctions in the SET-domain gene collections in the unicellular and in the multicellular (filamentous) relatives; diversification of SET-domain gene families has increased further with the expansion and elaboration of multicellularity in animal and plant systems. We found several ascomycota-specific SET-domain gene groups; each was unique to either Saccharomycotina or Pezizomycotina fungi. Our analysis revealed that the numbers and types of SET-domain genes in the Saccharomycotina did not reflect the habitats, pathogenicity, mechanisms of sexuality, or the ability to undergo morphogenic transformations. However, novel genes have appeared for functions associated with the transition to multicellularity. Descendents of most of the SET-domain gene families found in the filamentous fungi could be traced in the genomes of extant animals and plants, albeit as more complex structural forms.
Conclusion: SET-domain genes found in the filamentous species but absent from the unicellular sister group reflect two alternative evolutionary events: deletion from the yeast genomes or appearance of novel structures in filamentous fungal groups. There were no Ascomycota-specific SET-domain gene families (i.e., absent from animal and plant genomes); however, plants and animals share SET-domain gene subfamilies that do not exist in the fungi. Phylogenetic and gene-structure analyses defined several animal and plant SET-domain genes as sister groups while those of fungal origin were basal to them. Plants and animals also share SET-domain subfamilies that do not exist in fungi.
Figures


Similar articles
-
Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccharomycotina.BMC Genomics. 2007 Sep 17;8:325. doi: 10.1186/1471-2164-8-325. BMC Genomics. 2007. PMID: 17868481 Free PMC article.
-
Genome-scale phylogeny and contrasting modes of genome evolution in the fungal phylum Ascomycota.Sci Adv. 2020 Nov 4;6(45):eabd0079. doi: 10.1126/sciadv.abd0079. Print 2020 Nov. Sci Adv. 2020. PMID: 33148650 Free PMC article.
-
A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis.BMC Evol Biol. 2006 Nov 22;6:99. doi: 10.1186/1471-2148-6-99. BMC Evol Biol. 2006. PMID: 17121679 Free PMC article.
-
A complete inventory of fungal kinesins in representative filamentous ascomycetes.Fungal Genet Biol. 2003 Jun;39(1):1-15. doi: 10.1016/s1087-1845(03)00022-7. Fungal Genet Biol. 2003. PMID: 12742059 Review.
-
Self-eating to grow and kill: autophagy in filamentous ascomycetes.Appl Microbiol Biotechnol. 2013 Nov;97(21):9277-90. doi: 10.1007/s00253-013-5221-2. Appl Microbiol Biotechnol. 2013. PMID: 24077722 Review.
Cited by
-
Polycomb Repression without Bristles: Facultative Heterochromatin and Genome Stability in Fungi.Genes (Basel). 2020 Jun 9;11(6):638. doi: 10.3390/genes11060638. Genes (Basel). 2020. PMID: 32527036 Free PMC article. Review.
-
Characterization of the Polycomb-Group Mark H3K27me3 in Unicellular Algae.Front Plant Sci. 2017 Apr 26;8:607. doi: 10.3389/fpls.2017.00607. eCollection 2017. Front Plant Sci. 2017. PMID: 28491069 Free PMC article.
-
Deep evolutionary origins of neurobiology: Turning the essence of 'neural' upside-down.Commun Integr Biol. 2009;2(1):60-5. doi: 10.4161/cib.2.1.7620. Commun Integr Biol. 2009. PMID: 19513267 Free PMC article.
-
Plant neurobiology: from sensory biology, via plant communication, to social plant behavior.Cogn Process. 2009 Feb;10 Suppl 1:S3-7. doi: 10.1007/s10339-008-0239-6. Epub 2008 Nov 8. Cogn Process. 2009. PMID: 18998182 Review.
-
Histone methyltransferase ATX1 dynamically regulates fiber secondary cell wall biosynthesis in Arabidopsis inflorescence stem.Nucleic Acids Res. 2021 Jan 11;49(1):190-205. doi: 10.1093/nar/gkaa1191. Nucleic Acids Res. 2021. PMID: 33332564 Free PMC article.
References
-
- Alvarez-Venegas R, Sadder M, Tikhonov A, Avramova Z. Origin of the Bacterial SET Domain Genes: Vertical or Horizontal? Mol Biol Evol. 2007;24:482–497. - PubMed
-
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. - PubMed
-
- Jenuwein T, Allis CD. Translating the Histone Code. Science. 2001;298:1074. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

