Physiological and biochemical defects in carboxyl-terminal mutants of mitochondrial DNA helicase
- PMID: 18593709
- PMCID: PMC2527111
- DOI: 10.1074/jbc.M803674200
Physiological and biochemical defects in carboxyl-terminal mutants of mitochondrial DNA helicase
Abstract
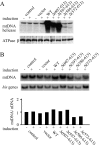
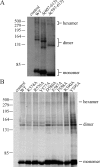
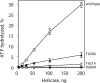
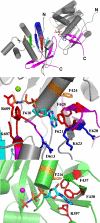
Mitochondrial DNA helicase, also called Twinkle, is essential for mtDNA maintenance. Its helicase domain shares high homology with helicases from superfamily 4. Structural analyses of helicases from this family indicate that carboxyl-terminal residues contribute to NTP hydrolysis required for translocation and DNA unwinding, yet genetic and biochemical information is very limited. Here, we evaluate the effects of overexpression in Drosophila cell culture of variants carrying a series of deletion and alanine substitution mutations in the carboxyl terminus and identify critical residues between amino acids 572 and 596 of the 613 amino acid polypeptide that are essential for mitochondrial DNA helicase function in vivo. Likewise, amino acid substitution mutants K574A, R576A, Y577A, F588A, and F595A show dose-dependent dominant-negative phenotypes. Arg-576 and Phe-588 are analogous to the arginine finger and base stack of other helicases, including the bacteriophage T7 gene 4 protein and bacterial DnaB helicase, respectively. We show here that representative human recombinant proteins that are analogous to the alanine substitution mutants exhibit defects in nucleotide hydrolysis. Our findings may be applicable to understand the role of the carboxyl-terminal region in superfamily 4 DNA helicases in general.
Figures


Similar articles
-
Autoregulation of the real-time kinetics of the human mitochondrial replicative helicase.Nat Commun. 2025 Jul 1;16(1):5460. doi: 10.1038/s41467-025-60289-0. Nat Commun. 2025. PMID: 40592829 Free PMC article.
-
Functional importance of the conserved N-terminal domain of the mitochondrial replicative DNA helicase.Biochim Biophys Acta. 2009 May;1787(5):290-5. doi: 10.1016/j.bbabio.2008.11.005. Epub 2008 Nov 21. Biochim Biophys Acta. 2009. PMID: 19063859 Free PMC article.
-
A Bacteriophage-Derived Primase-Helicase Orchestrates Plant Organellar DNA Replication.Physiol Plant. 2025 Jul-Aug;177(4):e70379. doi: 10.1111/ppl.70379. Physiol Plant. 2025. PMID: 40620047 Free PMC article.
-
Omega-3 fatty acids for depression in adults.Cochrane Database Syst Rev. 2015 Nov 5;2015(11):CD004692. doi: 10.1002/14651858.CD004692.pub4. Cochrane Database Syst Rev. 2015. Update in: Cochrane Database Syst Rev. 2021 Nov 24;11:CD004692. doi: 10.1002/14651858.CD004692.pub5. PMID: 26537796 Free PMC article. Updated.
-
Inhaled mannitol for cystic fibrosis.Cochrane Database Syst Rev. 2018 Feb 9;2(2):CD008649. doi: 10.1002/14651858.CD008649.pub3. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2020 May 1;5:CD008649. doi: 10.1002/14651858.CD008649.pub4. PMID: 29424930 Free PMC article. Updated.
Cited by
-
DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase.J Biol Chem. 2014 Oct 24;289(43):29975-93. doi: 10.1074/jbc.M114.567073. Epub 2014 Sep 5. J Biol Chem. 2014. PMID: 25193669 Free PMC article.
-
TWINKLE and Other Human Mitochondrial DNA Helicases: Structure, Function and Disease.Genes (Basel). 2020 Apr 9;11(4):408. doi: 10.3390/genes11040408. Genes (Basel). 2020. PMID: 32283748 Free PMC article. Review.
-
Inherited mitochondrial genomic instability and chemical exposures.Toxicology. 2017 Nov 1;391:75-83. doi: 10.1016/j.tox.2017.07.014. Epub 2017 Jul 26. Toxicology. 2017. PMID: 28756246 Free PMC article. Review.
-
Mitochondrial DNA maintenance: an appraisal.Mol Cell Biochem. 2015 Nov;409(1-2):283-305. doi: 10.1007/s11010-015-2532-x. Epub 2015 Aug 19. Mol Cell Biochem. 2015. PMID: 26286847 Review.
-
Ataxia.Neurol Clin. 2015 Feb;33(1):225-48. doi: 10.1016/j.ncl.2014.09.004. Neurol Clin. 2015. PMID: 25432731 Free PMC article. Review.
References
-
- Spelbrink, J. N., Li, F. Y., Tiranti, V., Nikali, K., Yuan, Q. P., Tariq, M., Wanrooij, S., Garrido, N., Comi, G., Morandi, L., Santoro, L., Toscano, A., Fabrizi, G. M., Somer, H., Croxen, R., Beeson, D., Poulton, J., Suomalainen, A., Jacobs, H. T., Zeviani, M., and Larsson, C. (2001) Nat. Genet. 28 223-231 - PubMed
-
- Sarzi, E., Goffart, S., Serre, V., Chretien, D., Slama, A., Munnich, A., Spelbrink, J. N., and Rotig, A. (2007) Ann. Neurol. 62579 -587 - PubMed
-
- Hakonen, A. H., Isohanni, P., Paetau, A., Herva, R., Suomalainen, A., and Lonnqvist, T. (2007) Brain 1303032 -3040 - PubMed
-
- Spinazzola, A., and Zeviani, M. (2005) Gene (Amst.) 354162 -168 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

