Identification of Plasmodium falciparum RhopH3 protein peptides that specifically bind to erythrocytes and inhibit merozoite invasion
- PMID: 18593818
- PMCID: PMC2548369
- DOI: 10.1110/ps.035923.108
Identification of Plasmodium falciparum RhopH3 protein peptides that specifically bind to erythrocytes and inhibit merozoite invasion
Abstract
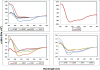
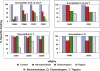
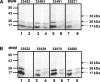
The identification of sequences involved in binding to erythrocytes is an important step for understanding the molecular basis of merozoite-erythrocyte interactions that take place during invasion of the Plasmodium falciparum malaria parasite into host cells. Several molecules located in the apical organelles (micronemes, rhoptry, dense granules) of the invasive-stage parasite are essential for erythrocyte recognition, invasion, and establishment of the nascent parasitophorous vacuole. Particularly, it has been demonstrated that rhoptry proteins play an important role in binding to erythrocyte surface receptors, among which is the PfRhopH3 protein, which triggers important immune responses in patients from endemic regions. It has also been reported that anti-RhopH3 antibodies inhibit in vitro invasion of erythrocytes, further supporting its direct involvement in erythrocyte invasion processes. In this study, PfRhopH3 consecutive peptides were synthesized and tested in erythrocyte binding assays for identifying those regions mediating binding to erythrocytes. Fourteen PfRhopH3 peptides presenting high specific binding activity were found, whose bindings were saturable and presented nanomolar dissociation constants. These high-activity binding peptides (HABPs) were characterized by having alpha-helical structural elements, as determined by circular dichroism, and having receptors of a possible sialic acid-dependent and/or glycoprotein-dependent nature, as evidenced in enzyme-treated erythrocyte binding assays and further corroborated by cross-linking assay results. Furthermore, these HABPs inhibited merozoite in vitro invasion of normal erythrocytes at 200 microM by up to 60% and 90%, suggesting that some RhopH3 protein regions are involved in the P. falciparum erythrocyte invasion.
Figures



Similar articles
-
Plasmodium falciparum rhoptry neck protein 5 peptides bind to human red blood cells and inhibit parasite invasion.Peptides. 2014 Mar;53:210-7. doi: 10.1016/j.peptides.2013.07.028. Epub 2013 Aug 8. Peptides. 2014. PMID: 23932940
-
Identification of a potent combination of key Plasmodium falciparum merozoite antigens that elicit strain-transcending parasite-neutralizing antibodies.Infect Immun. 2013 Feb;81(2):441-51. doi: 10.1128/IAI.01107-12. Epub 2012 Nov 26. Infect Immun. 2013. PMID: 23184525 Free PMC article.
-
PfRON3 is an erythrocyte-binding protein and a potential blood-stage vaccine candidate antigen.Malar J. 2014 Dec 12;13:490. doi: 10.1186/1475-2875-13-490. Malar J. 2014. PMID: 25495792 Free PMC article.
-
Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria.FEMS Microbiol Rev. 2016 May;40(3):343-72. doi: 10.1093/femsre/fuw001. Epub 2016 Jan 31. FEMS Microbiol Rev. 2016. PMID: 26833236 Free PMC article. Review.
-
Invasion of erythrocytes by malaria parasites: a cellular and molecular overview.Annu Rev Microbiol. 1986;40:451-77. doi: 10.1146/annurev.mi.40.100186.002315. Annu Rev Microbiol. 1986. PMID: 3535649 Review.
Cited by
-
Family members stick together: multi-protein complexes of malaria parasites.Med Microbiol Immunol. 2010 Aug;199(3):209-26. doi: 10.1007/s00430-010-0157-y. Epub 2010 Apr 24. Med Microbiol Immunol. 2010. PMID: 20419315 Review.
-
A large scale Plasmodium vivax- Saimiri boliviensis trophozoite-schizont transition proteome.PLoS One. 2017 Aug 22;12(8):e0182561. doi: 10.1371/journal.pone.0182561. eCollection 2017. PLoS One. 2017. PMID: 28829774 Free PMC article.
-
Long- and short-term selective forces on malaria parasite genomes.PLoS Genet. 2010 Sep 9;6(9):e1001099. doi: 10.1371/journal.pgen.1001099. PLoS Genet. 2010. PMID: 20838588 Free PMC article.
-
Human Cyclophilin B forms part of a multi-protein complex during erythrocyte invasion by Plasmodium falciparum.Nat Commun. 2017 Nov 16;8(1):1548. doi: 10.1038/s41467-017-01638-6. Nat Commun. 2017. PMID: 29146974 Free PMC article.
-
Purifying and Characterizing Bacterially Expressed Soluble Lactate Dehydrogenase from Plasmodium knowlesi for the Development of Anti-Malarial Drugs.Molecules. 2021 Nov 1;26(21):6625. doi: 10.3390/molecules26216625. Molecules. 2021. PMID: 34771034 Free PMC article.
References
-
- Anthony, R.N., Yang, J., Krall, J.A., Sam-Yellowe, T.Y. Sequence analysis of the Rhop-3 gene of Plasmodium yoelii . J. Eukaryot. Microbiol. 2000;47:319–322. - PubMed
-
- Bannister, L., Mitchell, G. The ins, outs and roundabouts of malaria. Trends Parasitol. 2003;19:209–213. - PubMed
-
- Breman, J.G., Alilio, M.S., Mills, A. Conquering the intolerable burden of malaria: What's new, what's needed: A summary. Am. J. Trop. Med. Hyg. 2004;71:1–15. - PubMed
-
- Brown, H.J., Coppel, R.L. Primary structure of a Plasmodium falciparum rhoptry antigen. Mol. Biochem. Parasitol. 1991;49:99–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

