The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock
- PMID: 18593878
- PMCID: PMC2492663
- DOI: 10.1101/gad.1682708
The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock
Abstract
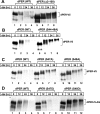
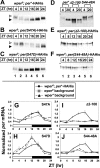
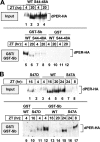
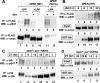
A common feature of animal circadian clocks is the progressive phosphorylation of PERIOD (PER) proteins, which is highly dependent on casein kinase Idelta/epsilon (CKIdelta/epsilon; termed DOUBLETIME [DBT] in Drosophila) and ultimately leads to the rapid degradation of hyperphosphorylated isoforms via a mechanism involving the F-box protein, beta-TrCP (SLIMB in Drosophila). Here we use the Drosophila melanogaster model system, and show that a key step in controlling the speed of the clock is phosphorylation of an N-terminal Ser (S47) by DBT, which collaborates with other nearby phosphorylated residues to generate a high-affinity atypical SLIMB-binding site on PER. DBT-dependent increases in the phospho-occupancy of S47 are temporally gated, dependent on the centrally located DBT docking site on PER and partially counterbalanced by protein phosphatase activity. We propose that the gradual DBT-mediated phosphorylation of a nonconsensus SLIMB-binding site establishes a temporal threshold for when in a daily cycle the majority of PER proteins are tagged for rapid degradation. Surprisingly, most of the hyperphosphorylation is unrelated to direct effects on PER stability. We also use mass spectrometry to map phosphorylation sites on PER, leading to the identification of a number of "phospho-clusters" that explain several of the classic per mutants.
Figures


Comment in
-
PERspective on PER phosphorylation.Genes Dev. 2008 Jul 1;22(13):1737-40. doi: 10.1101/gad.1696408. Genes Dev. 2008. PMID: 18593875 Free PMC article.
Similar articles
-
PERspective on PER phosphorylation.Genes Dev. 2008 Jul 1;22(13):1737-40. doi: 10.1101/gad.1696408. Genes Dev. 2008. PMID: 18593875 Free PMC article.
-
Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime.Nature. 2002 Dec 12;420(6916):673-8. doi: 10.1038/nature01272. Epub 2002 Nov 20. Nature. 2002. PMID: 12442174
-
A Doubletime Nuclear Localization Signal Mediates an Interaction with Bride of Doubletime to Promote Circadian Function.J Biol Rhythms. 2015 Aug;30(4):302-17. doi: 10.1177/0748730415588189. Epub 2015 Jun 16. J Biol Rhythms. 2015. PMID: 26082158 Free PMC article.
-
A PER/TIM/DBT interval timer for Drosophila's circadian clock.Cold Spring Harb Symp Quant Biol. 2007;72:69-74. doi: 10.1101/sqb.2007.72.034. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419263 Review.
-
The molecular control of circadian behavioral rhythms and their entrainment in Drosophila.Annu Rev Biochem. 1998;67:135-52. doi: 10.1146/annurev.biochem.67.1.135. Annu Rev Biochem. 1998. PMID: 9759485 Review.
Cited by
-
Circadian timekeeping and output mechanisms in animals.Curr Opin Neurobiol. 2013 Oct;23(5):724-31. doi: 10.1016/j.conb.2013.02.018. Epub 2013 May 31. Curr Opin Neurobiol. 2013. PMID: 23731779 Free PMC article. Review.
-
Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination.Science. 2015 Jan 30;347(6221):1257277. doi: 10.1126/science.1257277. Science. 2015. PMID: 25635104 Free PMC article.
-
Death of a Protein: The Role of E3 Ubiquitin Ligases in Circadian Rhythms of Mice and Flies.Int J Mol Sci. 2022 Sep 12;23(18):10569. doi: 10.3390/ijms231810569. Int J Mol Sci. 2022. PMID: 36142478 Free PMC article. Review.
-
PERspective on PER phosphorylation.Genes Dev. 2008 Jul 1;22(13):1737-40. doi: 10.1101/gad.1696408. Genes Dev. 2008. PMID: 18593875 Free PMC article.
-
Kinetics of doubletime kinase-dependent degradation of the Drosophila period protein.J Biol Chem. 2011 Aug 5;286(31):27654-62. doi: 10.1074/jbc.M111.243618. Epub 2011 Jun 9. J Biol Chem. 2011. PMID: 21659538 Free PMC article.
References
-
- Akten B., Jauch E., Genova G.K., Kim E.Y., Edery I., Raabe T., Jackson F.R. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 2003;6:251–257. - PubMed
-
- Bae K., Edery I. Regulating a circadian clock’s period, phase and amplitude by phosphorylation: Insights from Drosophila. J. Biochem. 2006;140:609–617. - PubMed
-
- Baylies M.K., Bargiello T.A., Jackson F.R., Young M.W. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. - PubMed
-
- Baylies M.K., Vosshall L.B., Sehgal A., Young M.W. New short period mutations of the Drosophila clock gene per. Neuron. 1992;9:575–581. - PubMed
-
- Chang D.C., Reppert S.M. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr. Biol. 2003;13:758–762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
