Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program
- PMID: 18593883
- PMCID: PMC2492669
- DOI: 10.1101/gad.477908
Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program
Abstract
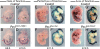
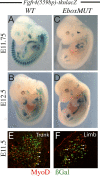
Pax3/7-dependent stem cells play an essential role in skeletal muscle development. We now show that Fgfr4 lies genetically downstream from Pax3 and is a direct target. In chromatin immunoprecipitation (ChIP)-on-chip experiments, Pax3 binds to a sequence 3' of the Fgfr4 gene that directs Pax3-dependent expression at sites of myogenesis in transgenic mouse embryos. The activity of this regulatory element is also partially dependent on E-boxes, targets of the myogenic regulatory factors, which are expressed as progenitor cells enter the myogenic program. Other FGF signaling components, notably Sprouty1, are also regulated by Pax3. In vivo manipulation of Sprouty expression reveals that FGF signaling affects the balance between Pax-positive progenitor cells and committed myoblasts. These results provide new insight into the Pax-initiated regulatory network that modulates stem cell maintenance versus tissue differentiation.
Figures


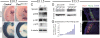
Similar articles
-
Regulation of skeletal muscle stem cell behavior by Pax3 and Pax7.Cold Spring Harb Symp Quant Biol. 2008;73:307-15. doi: 10.1101/sqb.2008.73.006. Epub 2008 Nov 6. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19022756 Review.
-
Expression patterns of FSHD-causing DUX4 and myogenic transcription factors PAX3 and PAX7 are spatially distinct in differentiating human stem cell cultures.Skelet Muscle. 2017 Jun 21;7(1):13. doi: 10.1186/s13395-017-0130-1. Skelet Muscle. 2017. PMID: 28637492 Free PMC article.
-
Dll4 and PDGF-BB convert committed skeletal myoblasts to pericytes without erasing their myogenic memory.Dev Cell. 2013 Mar 25;24(6):586-99. doi: 10.1016/j.devcel.2013.01.022. Epub 2013 Mar 7. Dev Cell. 2013. PMID: 23477786
-
A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb.Genes Dev. 2006 Sep 1;20(17):2450-64. doi: 10.1101/gad.382806. Genes Dev. 2006. PMID: 16951257 Free PMC article.
-
The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions.Annu Rev Cell Dev Biol. 2007;23:645-73. doi: 10.1146/annurev.cellbio.23.090506.123438. Annu Rev Cell Dev Biol. 2007. PMID: 17506689 Review.
Cited by
-
Spry1 and spry2 are essential for development of the temporomandibular joint.J Dent Res. 2012 Apr;91(4):387-93. doi: 10.1177/0022034512438401. Epub 2012 Feb 10. J Dent Res. 2012. PMID: 22328578 Free PMC article.
-
Loss of adult skeletal muscle stem cells drives age-related neuromuscular junction degeneration.Elife. 2017 Jun 6;6:e26464. doi: 10.7554/eLife.26464. Elife. 2017. PMID: 28583253 Free PMC article.
-
RIP-Seq of EZH2 Identifies TCONS-00036665 as a Regulator of Myogenesis in Pigs.Front Cell Dev Biol. 2021 Jan 12;8:618617. doi: 10.3389/fcell.2020.618617. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33511127 Free PMC article.
-
Myosin heavy chain-embryonic regulates skeletal muscle differentiation during mammalian development.Development. 2020 Apr 6;147(7):dev184507. doi: 10.1242/dev.184507. Development. 2020. PMID: 32094117 Free PMC article.
-
The FGFRL1 receptor is shed from cell membranes, binds fibroblast growth factors (FGFs), and antagonizes FGF signaling in Xenopus embryos.J Biol Chem. 2010 Jan 15;285(3):2193-202. doi: 10.1074/jbc.M109.058248. Epub 2009 Nov 17. J Biol Chem. 2010. PMID: 19920134 Free PMC article.
References
-
- Ben-Yair R., Kalcheim C. Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development. 2005;132:689–701. - PubMed
-
- Bladt F., Riethmacher D., Isenmann S., Aguzzi A., Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. - PubMed
-
- Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev. 2006;16:525–532. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
