Triptolide-induced transcriptional arrest is associated with changes in nuclear substructure
- PMID: 18593926
- PMCID: PMC2587069
- DOI: 10.1158/0008-5472.CAN-07-6207
Triptolide-induced transcriptional arrest is associated with changes in nuclear substructure
Abstract
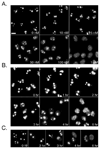
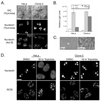
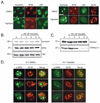
Triptolide, an active component of the medicinal herb lei gong teng, is a potent anticancer and anti-inflammatory therapeutic. It potently inhibits nuclear factor-kappaB transcriptional activation after DNA binding, although a precise mechanism is as yet unknown. Here, we report that triptolide also induces distinct nuclear substructural changes in HeLa cells. These changes in the nucleolus and nuclear speckles are reversible and dependent on both time and concentration. Furthermore, nuclear changes occurred within hours of triptolide treatment and were calcium and caspase independent. Rounding of nuclear speckles, an indication of transcriptional arrest, was evident and was associated with a decrease in RNA polymerase II (RNA Pol II) COOH-terminal domain Ser(2) phosphorylation. Additionally, the nucleolus disassembled and RNA Pol I activity declined after RNA Pol II inhibition. We therefore conclude that triptolide causes global transcriptional arrest as evidenced by inactivity of RNA Pol I and II and the subsequent alteration in nuclear substructure.
Figures



Similar articles
-
Triptolide (TPL) inhibits global transcription by inducing proteasome-dependent degradation of RNA polymerase II (Pol II).PLoS One. 2011;6(9):e23993. doi: 10.1371/journal.pone.0023993. Epub 2011 Sep 13. PLoS One. 2011. PMID: 21931633 Free PMC article.
-
Mechanism of triptolide-induced apoptosis: Effect on caspase activation and Bid cleavage and essentiality of the hydroxyl group of triptolide.J Mol Med (Berl). 2006 May;84(5):405-15. doi: 10.1007/s00109-005-0022-4. Epub 2005 Dec 30. J Mol Med (Berl). 2006. PMID: 16385419
-
Triptolide inhibits tumor promoter-induced uPAR expression via blocking NF-kappaB signaling in human gastric AGS cells.Anticancer Res. 2007 Sep-Oct;27(5A):3411-7. Anticancer Res. 2007. PMID: 17970088
-
Triptolide Induces Cell Killing in Multidrug-Resistant Tumor Cells via CDK7/RPB1 Rather than XPB or p44.Mol Cancer Ther. 2016 Jul;15(7):1495-503. doi: 10.1158/1535-7163.MCT-15-0753. Epub 2016 Mar 29. Mol Cancer Ther. 2016. PMID: 27197304
-
Broad targeting of triptolide to resistance and sensitization for cancer therapy.Biomed Pharmacother. 2018 Aug;104:771-780. doi: 10.1016/j.biopha.2018.05.088. Epub 2018 May 29. Biomed Pharmacother. 2018. PMID: 29807227 Review.
Cited by
-
Antagonist effect of triptolide on AKT activation by truncated retinoid X receptor-alpha.PLoS One. 2012;7(4):e35722. doi: 10.1371/journal.pone.0035722. Epub 2012 Apr 24. PLoS One. 2012. PMID: 22545132 Free PMC article.
-
Increased accumulation of hypoxia-inducible factor-1α with reduced transcriptional activity mediates the antitumor effect of triptolide.Mol Cancer. 2010 Oct 11;9:268. doi: 10.1186/1476-4598-9-268. Mol Cancer. 2010. PMID: 20932347 Free PMC article.
-
Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency.Cancer Cell. 2012 Apr 17;21(4):547-62. doi: 10.1016/j.ccr.2012.02.028. Cancer Cell. 2012. PMID: 22516262 Free PMC article.
-
Seminiferous epithelium damage after short period of busulphan treatment in adult rats and vitamin B12 efficacy in the recovery of spermatogonial germ cells.Int J Exp Pathol. 2016 Aug;97(4):317-328. doi: 10.1111/iep.12195. Epub 2016 Aug 31. Int J Exp Pathol. 2016. PMID: 27578607 Free PMC article.
-
A Bibliometric Analysis of Triptolide and the Recent Advances in Treating Non-Small Cell Lung Cancer.Front Pharmacol. 2022 May 30;13:878726. doi: 10.3389/fphar.2022.878726. eCollection 2022. Front Pharmacol. 2022. PMID: 35721205 Free PMC article. Review.
References
-
- Wu Y, Cui J, Bao X, et al. Triptolide attenuates oxidative stress, NF-kappaB activation and multiple cytokine gene expression in murine peritoneal macrophage. Int J Mol Med. 2006;17:141–150. - PubMed
-
- Zhao G, Vaszar LT, Qiu D, Shi L, Kao PN. Anti-inflammatory effects of triptolide in human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2000;279:L958–L966. - PubMed
-
- Choi YJ, Kim TG, Kim YH, et al. Immunosuppressant PG490 (triptolide) induces apoptosis through the activation of caspase-3 and down-regulation of XIAP in U937 cells. Biochem Pharmacol. 2003;66:273–280. - PubMed
-
- Wan CK, Wang C, Cheung HY, Yang M, Fong WF. Triptolide induces Bcl-2 cleavage and mitochondria dependent apoptosis in p53-deficient HL-60 cells. Cancer Lett. 2006;241:31–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

