Gene network dynamics controlling keratinocyte migration
- PMID: 18594517
- PMCID: PMC2516358
- DOI: 10.1038/msb.2008.36
Gene network dynamics controlling keratinocyte migration
Abstract
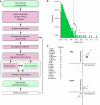
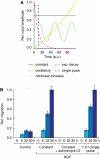
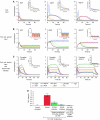
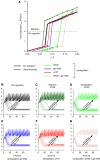
Translation of large-scale data into a coherent model that allows one to simulate, predict and control cellular behavior is far from being resolved. Assuming that long-term cellular behavior is reflected in the gene expression kinetics, we infer a dynamic gene regulatory network from time-series measurements of DNA microarray data of hepatocyte growth factor-induced migration of primary human keratinocytes. Transferring the obtained interactions to the level of signaling pathways, we predict in silico and verify in vitro the necessary and sufficient time-ordered events that control migration. We show that pulse-like activation of the proto-oncogene receptor Met triggers a responsive state, whereas time sequential activation of EGF-R is required to initiate and maintain migration. Context information for enhancing, delaying or stopping migration is provided by the activity of the protein kinase A signaling pathway. Our study reveals the complex orchestration of multiple pathways controlling cell migration.
Figures

Comment in
-
When microarrays Met epidermal-cell migration.Mol Syst Biol. 2008;4:200. doi: 10.1038/msb.2008.41. Epub 2008 Jul 1. Mol Syst Biol. 2008. PMID: 18594518 Free PMC article. No abstract available.
Similar articles
-
Boolean approach to signalling pathway modelling in HGF-induced keratinocyte migration.Bioinformatics. 2012 Sep 15;28(18):i495-i501. doi: 10.1093/bioinformatics/bts410. Bioinformatics. 2012. PMID: 22962472 Free PMC article.
-
Directed random walks and constraint programming reveal active pathways in hepatocyte growth factor signaling.FEBS J. 2016 Jan;283(2):350-60. doi: 10.1111/febs.13580. Epub 2015 Nov 26. FEBS J. 2016. PMID: 26518250
-
Role of ERK and JNK pathways in regulating cell motility and matrix metalloproteinase 9 production in growth factor-stimulated human epidermal keratinocytes.J Cell Physiol. 1999 Aug;180(2):271-84. doi: 10.1002/(SICI)1097-4652(199908)180:2<271::AID-JCP15>3.0.CO;2-D. J Cell Physiol. 1999. PMID: 10395297
-
EGFR is dispensable for c-Met-mediated proliferation and survival activities in mouse adult liver oval cells.Cell Signal. 2012 Feb;24(2):505-513. doi: 10.1016/j.cellsig.2011.09.031. Epub 2011 Oct 7. Cell Signal. 2012. PMID: 22001397
-
Hepatocyte growth factor/MET in cancer progression and biomarker discovery.Cancer Sci. 2017 Mar;108(3):296-307. doi: 10.1111/cas.13156. Cancer Sci. 2017. PMID: 28064454 Free PMC article. Review.
Cited by
-
When microarrays Met epidermal-cell migration.Mol Syst Biol. 2008;4:200. doi: 10.1038/msb.2008.41. Epub 2008 Jul 1. Mol Syst Biol. 2008. PMID: 18594518 Free PMC article. No abstract available.
-
From a traditional medicinal plant to a rational drug: understanding the clinically proven wound healing efficacy of birch bark extract.PLoS One. 2014 Jan 22;9(1):e86147. doi: 10.1371/journal.pone.0086147. eCollection 2014. PLoS One. 2014. PMID: 24465925 Free PMC article.
-
Emerging Roles for SSeCKS/Gravin/AKAP12 in the Control of Cell Proliferation, Cancer Malignancy, and Barriergenesis.Genes Cancer. 2010 Nov;1(11):1147-56. doi: 10.1177/1947601910392984. Genes Cancer. 2010. PMID: 21779438 Free PMC article.
-
Protein-bound uremic toxins induce tissue remodeling by targeting the EGF receptor.J Am Soc Nephrol. 2015 Feb;26(2):281-90. doi: 10.1681/ASN.2014010021. Epub 2014 Jul 10. J Am Soc Nephrol. 2015. PMID: 25012179 Free PMC article.
-
Stathmin regulates keratinocyte proliferation and migration during cutaneous regeneration.PLoS One. 2013 Sep 16;8(9):e75075. doi: 10.1371/journal.pone.0075075. eCollection 2013. PLoS One. 2013. PMID: 24066165 Free PMC article.
References
-
- Aldridge BB, Haller G, Sorger PK, Lauffenburger DA (2006) Direct lyapunov exponent analysis enables parametric study of transient signalling governing cell behaviour. IEE Proc Syst Biol 153: 425–432 - PubMed
-
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, Vaisman N, Segal E, Rechavi G, Alon U, Mills GB, Domany E, Yarden Y (2007) A module of negative feedback regulators defines growth factor signaling. Nat Genet 39: 503–512 - PubMed
-
- Asthagiri AR, Lauffenburger DA (2000) Bioengineering models of cell signaling. Annu Rev Biomed Eng 2: 31–53 - PubMed
-
- Banu N, Buda A, Chell S, Elder D, Moorghen M, Paraskeva C, Qualtrough D, Pignatelli M (2007) Inhibition of cox-2 with ns-398 decreases colon cancer cell motility through blocking epidermal growth factor receptor transactivation: possibilities for combination therapy. Cell Prolif 40: 768–779 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

