Bioinformatic analysis of the contribution of primer sequences to aptamer structures
- PMID: 18594898
- PMCID: PMC2671994
- DOI: 10.1007/s00239-008-9130-4
Bioinformatic analysis of the contribution of primer sequences to aptamer structures
Abstract
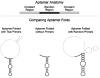
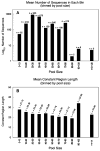
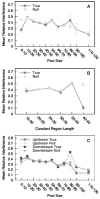
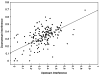
Aptamers are nucleic acid molecules selected in vitro to bind a particular ligand. While numerous experimental studies have examined the sequences, structures, and functions of individual aptamers, considerably fewer studies have applied bioinformatics approaches to try to infer more general principles from these individual studies. We have used a large Aptamer Database to parse the contributions of both random and constant regions to the secondary structures of more than 2000 aptamers. We find that the constant, primer-binding regions do not, in general, contribute significantly to aptamer structures. These results suggest that (a) binding function is not contributed to nor constrained by constant regions; (b) in consequence, the landscape of functional binding sequences is sparse but robust, favoring scenarios for short, functional nucleic acid sequences near origins; and (c) many pool designs for the selection of aptamers are likely to prove robust.
Figures

References
-
- Ancel L, Fontana W. Plasticity, modularity and evolvability in RNA in vitro. Exp Zool. 2000;288:242–283. - PubMed
-
- Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1417. - PubMed
-
- Coleman TM, Huang F. RNA-catalyzed thioester synthesis. Chem Biol. 2002;9:1227–1236. - PubMed
-
- Coleman TM, Huang F. Optimal random libraries for the isolation of catalytic RNA. RNA Biol. 2005;2:129–136. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

