Changes in population dynamics during long-term evolution of sabin type 1 poliovirus in an immunodeficient patient
- PMID: 18596089
- PMCID: PMC2546908
- DOI: 10.1128/JVI.00468-08
Changes in population dynamics during long-term evolution of sabin type 1 poliovirus in an immunodeficient patient
Abstract
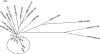
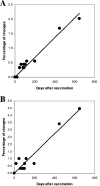
The evolution of the Sabin strain of type 1 poliovirus in a hypogammaglobulinemia patient for a period of 649 days is described. Twelve poliovirus isolates from sequential stool samples encompassing days 21 to 649 after vaccination with Sabin 1 were characterized in terms of their antigenic properties, virulence in transgenic mice, sensitivity for growth at high temperatures, and differences in nucleotide sequence from the Sabin 1 strain. Poliovirus isolates from the immunodeficient patient evolved gradually toward non-temperature-sensitive and neurovirulent phenotypes, accumulating mutations at key nucleotide positions that correlated with the observed reversion to biological properties typical of wild polioviruses. Analysis of plaque-purified viruses from stool samples revealed complex genetic and evolutionary relationships between the poliovirus strains. The generation of various coevolving genetic lineages incorporating different mutations was observed at early stages of virus excretion. The main driving force for genetic diversity appeared to be the selection of mutations at attenuation sites, particularly in the 5' noncoding region and the VP1 BC loop. Recombination between virus strains from the two main lineages was observed between days 63 and 88. Genetic heterogeneity among plaque-purified viruses at each time point seemed to decrease with time, and only viruses belonging to a unique genotypic lineage were seen from day 105 after vaccination. The relevance of vaccine-derived poliovirus strains for disease surveillance and future polio immunization policies is discussed in the context of the Global Polio Eradication Initiative.
Figures


References
-
- Alexander, L. N., J. F. Seward, T. A. Santibanez, M. A. Pallansch, O. M. Kew, D. R. Prevots, P. M. Strebel, J. Cono, M. Wharton, W. A. Orenstein, and R. W. Sutter. 2004. Vaccine policy changes and epidemiology of poliomyelitis in the United States. JAMA 2921696-1701. - PubMed
-
- Bellmunt, A., G. May, R. Zell, P. Pring-Akerblom, W. Verhagen, and A. Heim. 1999. Evolution of poliovirus type I during 5.5 years of prolonged enteral replication in an immunodeficient patient. Virology 265178-184. - PubMed
-
- Blomqvist, S., A. L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84573-580. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

