Analysis of natural sequence variation and covariation in human immunodeficiency virus type 1 integrase
- PMID: 18596095
- PMCID: PMC2546890
- DOI: 10.1128/JVI.01535-07
Analysis of natural sequence variation and covariation in human immunodeficiency virus type 1 integrase
Abstract
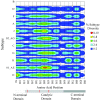
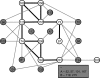
Human immunodeficiency virus type 1 (HIV-1) integrase inhibitors are in clinical trials, and raltegravir and elvitegravir are likely to be the first licensed drugs of this novel class of HIV antivirals. Understanding resistance to these inhibitors is important to maximize their efficacy. It has been shown that natural variation and covariation provide valuable insights into the development of resistance for established HIV inhibitors. Therefore, we have undertaken a study to fully characterize natural polymorphisms and amino acid covariation within an inhibitor-naïve sequence set spanning all defined HIV-1 subtypes. Inter- and intrasubtype variation was greatest in a 50-amino-acid segment of HIV-1 integrase incorporating the catalytic aspartic acid codon 116, suggesting that polymorphisms affect inhibitor binding and pathways to resistance. The critical mutations that determine the resistance pathways to raltegravir and elvitegravir (N155H, Q148K/R/H, and E92Q) were either rare or absent from the 1,165-sequence data set. However, 25 out of 41 mutations associated with integrase inhibitor resistance were present. These mutations were not subtype associated and were more prevalent in the subtypes that had been sampled frequently within the database. A novel modification of the Jaccard index was used to analyze amino acid covariation within HIV-1 integrase. A network of 10 covarying resistance-associated mutations was elucidated, along with a further 15 previously undescribed mutations that covaried with at least two of the resistance positions. The validation of covariation as a predictive tool will be dependent on monitoring the evolution of HIV-1 integrase under drug selection pressure.
Figures

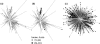

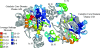
References
-
- Billich, A. 2003. S-1360 Shionogi-GlaxoSmithKline. Curr. Opin. Investig. Drugs 4206-209. - PubMed
-
- Brown, A. J., B. T. Korber, and J. H. Condra. 1999. Associations between amino acids in the evolution of HIV type 1 protease sequences under indinavir therapy. AIDS Res. Hum Retrovir. 15247-253. - PubMed
-
- D'Aquila, R. T., J. M. Schapiro, F. Brun-Vezinet, B. Clotet, B. Conway, L. M. Demeter, R. M. Grant, V. A. Johnson, D. R. Kuritzkes, C. Loveday, R. W. Shafer, and D. D. Richman. 2003. Drug resistance mutations in HIV-1. Top. HIV Med. 1192-96. - PubMed
-
- Embrey, M. W., J. S. Wai, T. W. Funk, C. F. Homnick, D. S. Perlow, S. D. Young, J. P. Vacca, D. J. Hazuda, P. J. Felock, K. A. Stillmock, M. V. Witmer, G. Moyer, W. A. Schleif, L. J. Gabryelski, L. Jin, I. W. Chen, J. D. Ellis, B. K. Wong, J. H. Lin, Y. M. Leonard, N. N. Tsou, and L. Zhuang. 2005. A series of 5-(5,6)-dihydrouracil substituted 8-hydroxy-[1,6]naphthyridine-7-carboxylic acid 4-fluorobenzylamide inhibitors of HIV-1 integrase and viral replication in cells. Bioorg. Med. Chem. Lett. 154550-4554. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

