Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
- PMID: 18596201
- PMCID: PMC2548357
- DOI: 10.1110/ps.036384.108
Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
Abstract
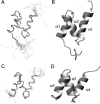
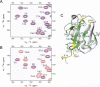
The Cbl proteins, RING-type E3 ubiquitin ligases, are responsible for ubiquitinating the activated tyrosine kinases and targeting them for degradation. Both c-Cbl and Cbl-b have a UBA (ubiquitin-associated) domain at their C-terminal ends, and these two UBA domains share a high sequence similarity (75%). However, only the UBA from Cbl-b, but not from c-Cbl, can bind ubiquitin (Ub). To understand the mechanism by which the UBA domains specifically interact with Ub with different affinities, we determined the solution NMR structures of these two UBA domains, cUBA from human c-Cbl and UBAb from Cbl-b. Their structures show that these two UBA domains share the same fold, a compact three-helix bundle, highly resembling the typical UBA fold. Chemical shift perturbation experiments reveal that the helix-1 and loop-1 of UBAb form a predominately hydrophobic surface for Ub binding. By comparing the Ub-interacting surface on UBAb and its counterpart on cUBA, we find that the hydrophobic patch on cUBA is interrupted by a negatively charged residue Glu12. Fluorescence titration data show that the Ala12Glu mutant of UBAb completely loses the ability to bind Ub, whereas the mutation disrupting the dimerization has no significant effect on Ub binding. This study provides structural and biochemical insights into the Ub binding specificities of the Cbl UBA domains, in which the hydrophobic surface distribution on the first helix plays crucial roles in their differential affinities for Ub binding. That is, the amino acid residue diversity in the helix-1 region, but not the dimerization, determines the abilities of various UBA domains binding with Ub.
Figures
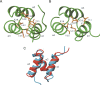


References
-
- Bartkiewicz, M., Houghton, A., Baron, R. Leucine zipper-mediated homodimerization of the adaptor protein c-Cbl. A role in c-Cbl's tyrosine phosphorylation and its association with epidermal growth factor receptor. J. Biol. Chem. 1999;274:30887–30895. - PubMed
-
- Blake, T.J., Langdon, W.Y. A rearrangement of the c-cbl proto-oncogene in HUT78 T-lymphoma cells results in a truncated protein. Oncogene. 1992;7:757–762. - PubMed
-
- Blake, T.J., Shapiro, M., Morse H.C., III, Langdon, W.Y. The sequences of the human and mouse c-cbl proto-oncogenes show v-cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene. 1991;6:653–657. - PubMed
-
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

