Spliceosomal snRNAs in the unicellular eukaryote Trichomonas vaginalis are structurally conserved but lack a 5'-cap structure
- PMID: 18596255
- PMCID: PMC2491460
- DOI: 10.1261/rna.1045408
Spliceosomal snRNAs in the unicellular eukaryote Trichomonas vaginalis are structurally conserved but lack a 5'-cap structure
Abstract
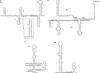
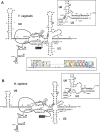
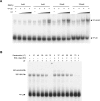
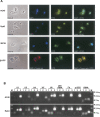
Few genes in the divergent eukaryote Trichomonas vaginalis have introns, despite the unusually large gene repertoire of this human-infective parasite. These introns are characterized by extended conserved regulatory motifs at the 5' and 3' boundaries, a feature shared with another divergent eukaryote, Giardia lamblia, but not with metazoan introns. This unusual characteristic of T. vaginalis introns led us to examine spliceosomal small nuclear RNAs (snRNAs) predicted to mediate splicing reactions via interaction with intron motifs. Here we identify T. vaginalis U1, U2, U4, U5, and U6 snRNAs, present predictions of their secondary structures, and provide evidence for interaction between the U2/U6 snRNA complex and a T. vaginalis intron. Structural models predict that T. vaginalis snRNAs contain conserved sequences and motifs similar to those found in other examined eukaryotes. These data indicate that mechanisms of intron recognition as well as coordination of the two catalytic steps of splicing have been conserved throughout eukaryotic evolution. Unexpectedly, we found that T. vaginalis spliceosomal snRNAs lack the 5' trimethylguanosine cap typical of snRNAs and appear to possess unmodified 5' ends. Despite the lack of a cap structure, U1, U2, U4, and U5 genes are transcribed by RNA polymerase II, whereas the U6 gene is transcribed by RNA polymerase III.
Figures


References
-
- Abrahamsen M.S., Templeton T.J., Enomoto S., Abrahante J.E., Zhu G., Lancto C.A., Deng M., Liu C., Widmer G., Tzipori S., et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum . Science. 2004;304:441–445. - PubMed
-
- Ares M., Jr, Weiser B. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog. Nucleic Acid Res. Mol. Biol. 1995;50:131–159. - PubMed
-
- Baldauf S.L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. - PubMed
-
- Braglia P., Percudani R., Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 2005;280:19551–19562. - PubMed
-
- Brow D.A., Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
