Identification of marker genes for differential diagnosis of chronic fatigue syndrome
- PMID: 18596870
- PMCID: PMC2442021
- DOI: 10.2119/2007-00059.Saiki
Identification of marker genes for differential diagnosis of chronic fatigue syndrome
Abstract
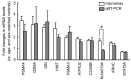
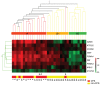
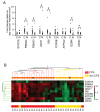
Chronic fatigue syndrome (CFS) is a clinically defined condition characterized by long-lasting disabling fatigue. Because of the unknown mechanism underlying this syndrome, there still is no specific biomarker for objective assessment of the pathological fatigue. We have compared gene expression profiles in peripheral blood between 11 drug-free patients with CFS and age- and sex-matched healthy subjects using a custom microarray carrying complementary DNA probes for 1,467 stress-responsive genes. We identified 12 genes whose mRNA levels were changed significantly in CFS patients. Of these 12 genes, quantitative real-time PCR validated the changes in 9 genes encoding granzyme in activated T or natural killer cells (GZMA), energy regulators (ATP5J2, COX5B, and DBI), proteasome subunits (PSMA3 and PSMA4), putative protein kinase c inhibitor (HINT ), GTPase (ARHC), and signal transducers and activators of transcription 5A (STAT5A). Next, we performed the same microarray analysis on 3 additional CFS patients and 20 other patients with the chief complaint of long-lasting fatigue related to other disorders (non-CFS patients) and found that the relative mRNA expression of 9 genes classified 79% (11/14) of CFS and 85% (17/20) of the non-CFS patients. Finally, real-time PCR measurements of the levels of the 9 involved mRNAs were done in another group of 18 CFS and 12 non-CFS patients. The expression pattern correctly classified 94% (17/18) of CFS and 92% (11/12) of non-CFS patients. Our results suggest that the defined gene cluster (9 genes) may be useful for detecting pathological responses in CFS patients and for differential diagnosis of this syndrome.
Figures

Similar articles
-
[Identification and application of marker genes for differential diagnosis of chronic fatigue syndrome].Nihon Rinsho. 2007 Jun;65(6):1029-33. Nihon Rinsho. 2007. PMID: 17561693 Review. Japanese.
-
Utility of the blood for gene expression profiling and biomarker discovery in chronic fatigue syndrome.Dis Markers. 2002;18(4):193-9. doi: 10.1155/2002/892374. Dis Markers. 2002. PMID: 12590173 Free PMC article.
-
MicroRNAs hsa-miR-99b, hsa-miR-330, hsa-miR-126 and hsa-miR-30c: Potential Diagnostic Biomarkers in Natural Killer (NK) Cells of Patients with Chronic Fatigue Syndrome (CFS)/ Myalgic Encephalomyelitis (ME).PLoS One. 2016 Mar 11;11(3):e0150904. doi: 10.1371/journal.pone.0150904. eCollection 2016. PLoS One. 2016. PMID: 26967895 Free PMC article.
-
Exercise responsive genes measured in peripheral blood of women with chronic fatigue syndrome and matched control subjects.BMC Physiol. 2005 Mar 24;5(1):5. doi: 10.1186/1472-6793-5-5. BMC Physiol. 2005. PMID: 15790422 Free PMC article.
-
Clinical Practice: Chronic fatigue syndrome.Eur J Pediatr. 2013 Oct;172(10):1293-8. doi: 10.1007/s00431-013-2058-8. Epub 2013 Jun 12. Eur J Pediatr. 2013. PMID: 23756916 Review.
Cited by
-
Review of the Quality Control Checks Performed by Current Genome-Wide and Targeted-Genome Association Studies on Myalgic Encephalomyelitis/Chronic Fatigue Syndrome.Front Pediatr. 2020 Jun 12;8:293. doi: 10.3389/fped.2020.00293. eCollection 2020. Front Pediatr. 2020. PMID: 32596192 Free PMC article. No abstract available.
-
Fatigue in rheumatic diseases.Eur J Rheumatol. 2015 Sep;2(3):109-113. doi: 10.5152/eurjrheum.2015.0029. Epub 2015 Sep 1. Eur J Rheumatol. 2015. PMID: 27708942 Free PMC article. Review.
-
Immune and hemorheological changes in chronic fatigue syndrome.J Transl Med. 2010 Jan 11;8:1. doi: 10.1186/1479-5876-8-1. J Transl Med. 2010. PMID: 20064266 Free PMC article.
-
mRNA transcripts as molecular biomarkers in medicine and nutrition.J Nutr Biochem. 2010 Aug;21(8):665-70. doi: 10.1016/j.jnutbio.2009.11.012. Epub 2010 Mar 20. J Nutr Biochem. 2010. PMID: 20303730 Free PMC article. Review.
-
Genetics and Gene Expression Involving Stress and Distress Pathways in Fibromyalgia with and without Comorbid Chronic Fatigue Syndrome.Pain Res Treat. 2012;2012:427869. doi: 10.1155/2012/427869. Epub 2011 Sep 29. Pain Res Treat. 2012. PMID: 22110941 Free PMC article.
References
-
- Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International chronic fatigue syndrome study group. Annals of Internal Medicine. 1994;121:953–9. - PubMed
-
- Powell R, Ren J, Lewith G, Barclay W, Holgate S, Almond J. Identification of novel expressed sequences, up-regulated in the leucocytes of chronic fatigue syndrome patients. Clin Exp Allergy. 2003;33:1450–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

