Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice
- PMID: 18596921
- PMCID: PMC2441855
- DOI: 10.1172/JCI34538
Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice
Abstract
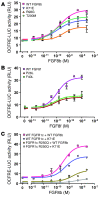
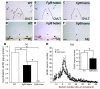
Idiopathic hypogonadotropic hypogonadism (IHH) with anosmia (Kallmann syndrome; KS) or with a normal sense of smell (normosmic IHH; nIHH) are heterogeneous genetic disorders associated with deficiency of gonadotropin-releasing hormone (GnRH). While loss-of-function mutations in FGF receptor 1 (FGFR1) cause human GnRH deficiency, to date no specific ligand for FGFR1 has been identified in GnRH neuron ontogeny. Using a candidate gene approach, we identified 6 missense mutations in FGF8 in IHH probands with variable olfactory phenotypes. These patients exhibited varied degrees of GnRH deficiency, including the rare adult-onset form of hypogonadotropic hypogonadism. Four mutations affected all 4 FGF8 splice isoforms (FGF8a, FGF8b, FGF8e, and FGF8f), while 2 mutations affected FGF8e and FGF8f isoforms only. The mutant FGF8b and FGF8f ligands exhibited decreased biological activity in vitro. Furthermore, mice homozygous for a hypomorphic Fgf8 allele lacked GnRH neurons in the hypothalamus, while heterozygous mice showed substantial decreases in the number of GnRH neurons and hypothalamic GnRH peptide concentration. In conclusion, we identified FGF8 as a gene implicated in GnRH deficiency in both humans and mice and demonstrated an exquisite sensitivity of GnRH neuron development to reductions in FGF8 signaling.
Figures


References
-
- Herbison, A.E. 2006. Physiology of the GnRH neuronal network. InKnobil and Neill’s physiology of reproduction. 3rd edition. J.D. Neill, editor. Academic Press. San Diego, California, USA. 1415–1482.
-
- Knobil E., Neill J.D., Johansson E.D. Influence of hypophysectomy, sham hypophysectomy and other surgical procedures on luteal function in the rhesus monkey. Endocrinology. 1968;82:410–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

