Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing
- PMID: 18597733
- PMCID: PMC2443835
- DOI: 10.1016/j.ajhg.2008.06.018
Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing
Abstract
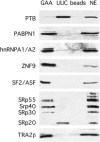
The intronic GAA repeat expansion in the frataxin (FXN) gene causes the hereditary neurodegenerative disorder Friedreich ataxia. Although it is generally believed that GAA repeats block transcription elongation, direct proof in eukaryotic systems is lacking. We tested in hybrid minigenes the effect of GAA and TTC repeats on nascent transcription and pre-mRNA processing. Unexpectedly, disease-causing GAA(100) repeats did not affect transcriptional elongation in a nuclear HeLa Run On assay, nor did they affect pre-mRNA transcript abundance. However, they did result in a complex defect in pre-mRNA processing. The insertion of GAA but not TTC repeats downstream of reporter exons resulted in their partial or complete exclusion from the mature mRNAs and in the generation of a variety of aberrant splicing products. This effect of GAA repeats was observed to be position and context dependent; their insertion at different distances from the reporter exons had a variable effect on splice-site selection. In addition, GAA repeats bind to a multitude of different splicing factors and induced the accumulation of an upstream pre-mRNA splicing intermediate, which is not turned over into mature mRNA. When embedded in the homologous frataxin minigene system, the GAA repeats did not affect the pre-mRNA transcript abundance but did significantly reduce the splicing efficiency of the first intron. These data indicate an association between GAA noncoding repeats and aberrant pre-mRNA processing because binding of transcribed GAA repeats to a multitude of trans-acting splicing factors can interfere with normal turnover of intronic RNA and thus lead to its degradation and a lower abundance of mature mRNA.
Figures






References
-
- Pandolfo M. The molecular basis of Friedreich ataxia. Adv. Exp. Med. Biol. 2002;516:99–118. - PubMed
-
- Harding A.E. Friedreich's ataxia: A clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain. 1981;104:589–620. - PubMed
-
- Pandolfo M., Koenig M. Friedreich's ataxia. In: Wells R.D., Warren S.T., editors. Genetic Instabilities and Hereditary Neuromuscular Diseases. Academic Press; San Diego, CA: 1998. pp. 373–398.
-
- Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A. Friedreich's ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. - PubMed
-
- Clark R.M., Dalgliesh G.L., Endres D., Gomez M., Taylor J., Bidichandani S.I. Expansion of GAA triplet repeats in the human genome: Unique origin of the FRDA mutation at the center of an Alu. Genomics. 2004;83:373–383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

